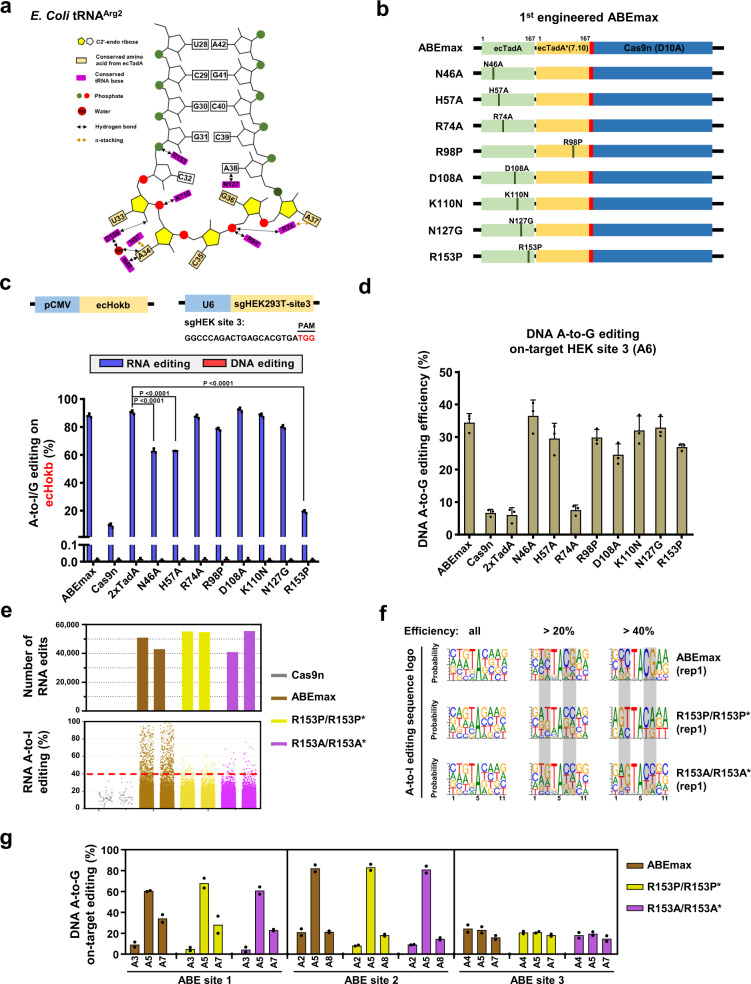
Fig. 1. Arginine 153 within TadA/TadA* is essential for specificity of ABEmax-induced RNA deamination.
a Summary of predicted E. coli TadA-tRNA interactions. A classical tRNAArg2 possessing CUACGAA and the potential interaction between the residues from TadA (purple) and tRNA loop nucleotides was shown. b Design for the engineered ABEmax (for the 1st round of engineering) with indicated amino acid substitutions. c Schematic overview of reporter assays for RNA editing. The upper diagram represents a gene Hokb cloned from E. coli that is driven by a CMV promoter (pCMV). An sgRNA targeting HEK-site 3 (sgHEK site 3) was constructed into a pGL3-U6 vector. The lower bar plot showed the percentage of A-to-I/G (A-to-G for genomic DNA test; A-to-I for RNA-generated cDNA test) conversion for Hokb gene in the reporter, which was co-transfected with sgHEK site 3 and ABEmax variants into HEK293T cells. After transfection for 72 h, cells were harvested for genomic DNA and RNA extraction and reverse transcription, followed by PCR amplification and deep sequencing. A adenine, I inosine, ABEmax codon optimized adenine base editor. Two-tailed t-test was performed for comparisons and P values were presented. d Genomic DNA was subjected to PCR amplification of DNA fragments containing HEK site 3 targeting sites for ABEmax variants, followed by deep sequencing. The A-to-G editing efficiency on adenine 6 (A6) within the sgRNA sequence was shown with triplicates. e The number (upper; by MuTect2) and efficiency distribution (lower in Jitter plot) of RNA A-to-I edits were presented for RNA-seq experiments in HEK293T cells that expressed Cas9n (rep1 and rep2), ABEmax (rep1 and rep2), ABEmax R153P/R153P* (rep1 and rep2), or ABEmax R153A/R153A* (rep1 and rep2) and the sgRNA targeting HEK site 8. Two independent replicates were shown. Each dot in the lower panel represents an edited adenine position in RNA. Non-transfected HEK293T transcriptome served as an endogenous editing control, and the RNA A-to-I edits overlapping with endogenous A-to-I edits were excluded. f Sequence logos centered by the edited adenine (A) from a single replicate of RNA-seq data in e (rep1 for each group) for edited adenines with indicated editing efficiencies (>40%, >20%, and all). The nucleotides around the edited A were shown (nine nucleotides in total). The changed sequence logos for ABEmax variants were highlighted in gray. g Bar plot showing the on-target DNA A-to-G editing efficiencies of ABEmax, ABEmax R153P/R153P*, and ABEmax R153A/R153A* with 3 sgRNAs targeting ABE site 1-3. The constructs for these three ABEmax variants were presented in Supplementary Fig. 4c. Data was generated from two independent replicates from deep sequencing analysis. The efficiencies for most highly edited adenines for each sgRNA on-target site within the editing window were reported with two replicates. Error bars represent s.d. Source data are available in the Source Data file.