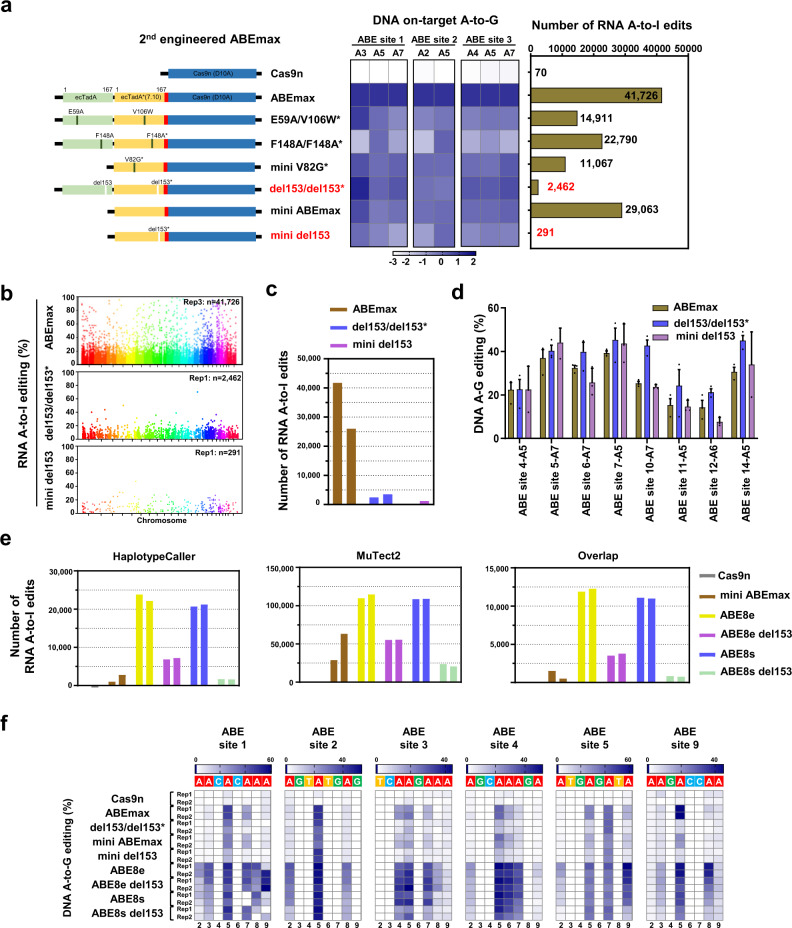
Fig. 2. Deletion of R153 from TadA/TadA* results in minimized RNA off-targeting activities.
a Left panel: design for the second round of engineered ABEmax variants with R153 deletions (del). Asterisk represents amino acid substitutions or deletions constructed in TadA*. mini ABEmax was fused by TadA* and Cas9n. R153 was deleted from TadA* to generate mini del153. Middle panel: heat maps showing DNA on-target A-to-G editing efficiency in HEK293T cells co-expressing ABEmax or its variants (as shown in left panel) with sgRNAs targeting ABE site 1, 2, and 3, respectively. Efficiencies were presented in heat map as log2(fold change) for each site; each box representing the DNA A-to-G editing efficiency normalized to the that observed with ABEmax for each on-target DNA sites (ABE site 1, 2, and 3). The efficiencies for edited adenines within the window (4-8 of the protospacers) of the on-target sites for these three sites were shown. Right panel: the number of RNA A-to-I edits (calculated by MuTect2) induced by engineered ABEmax variants was presented; ABEmax (rep3) and Cas9n (rep3) served as controls. Non-transfected HEK293T transcriptome served as a control, and the RNA A-to-I edits overlapping with endogenous A-to-I edits were excluded. b Manhattan plots showing the representative distributions of RNA A-to-I edits across all chromosomes induced by ABEmax (rep3), del153/dele153* (rep1), and mini del153 (rep1) with an sgRNA targeting ABE site 8 in HEK293T cells from RNA-seq data. c The diagram showing the number of RNA A-to-I edits induced by ABEmax (rep3 and rep4), del153/dele153* (rep1 and rep2), and mini del153 (rep1 and rep2) with an sgRNA targeting ABE site 8 in HEK293T cells from RNA-seq analysis. d Bar plots showing the DNA on-target A-to-G editing efficiencies of ABEmax, del153/dele153*, and mini del153 with 8 sgRNAs. The efficiencies for most highly edited adenines for each sgRNA on-target site within the editing window are reported, and error bars represent mean ± s.d. of three independent experiments. e The number of RNA edits calculated by HaplotypeCaller or MuTect2, or overlapped RNA edits (HaplotypeCaller and MuTect2) was presented for RNA-seq experiments in HEK293T cells that expressed Cas9n, mini ABEmax, ABE8e, ABE8e del153, ABE8s, ABE8s del153, and an sgRNA targeting HEK site 8. Two independent replicates were shown. f Heat maps showing DNA on-target A-to-G editing efficiencies of Cas9n, ABEmax, del153/dele153*, mini ABEmax, mini del153, ABE8e, ABE8e del153, ABE8s, and ABE8s del153 with six sgRNAs. Data are generated from two independent replicates. A-to-G editing efficiencies are shown in heat map format for the editing window 2–9. Numbering at the bottom of the heat map represents spacer position and “1” was calculated from the most PAM-distal nucleotide. Source data are available in the Source Data file.