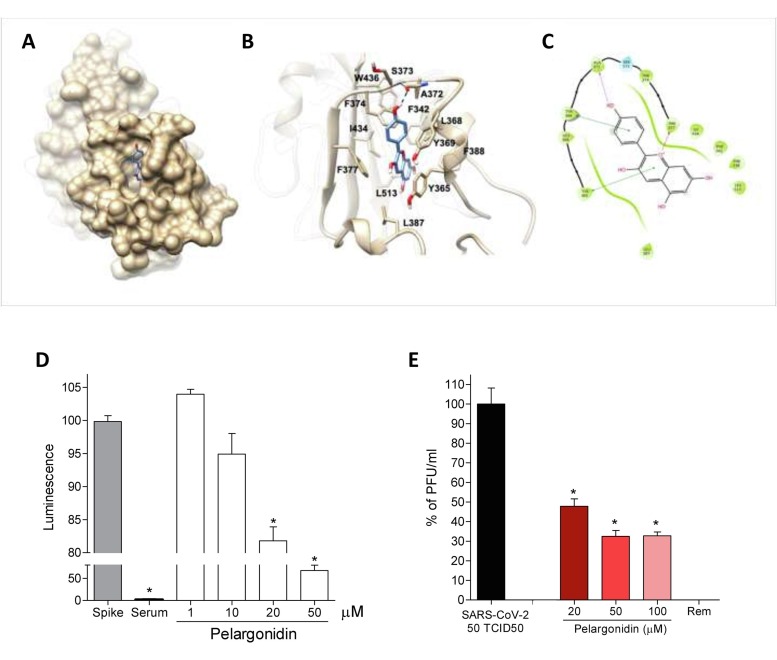
Fig. 8.
Pelargonidin inhibits binding of the SARS-Cov-2 virus on the host cells. (A) Hydrophobic FA binding pocket in a surface representation; (B) Cartoon representation of binding mode of pelargonidin to SARS-CoV-2 receptor. The ligand is represented as blue sticks, whereas the interacting residues of the receptor are shown in tan and labelled. Oxygen atoms are depicted in red and nitrogens in blue. The receptors are represented as tan ribbons. Hydrogens are omitted for the sake of clarity; (C) Diagram of Pelargonidin interaction. (D) SARS-CoV-2 Spike binding to immobilized ACE2; Luminescence was measured using a Fluo-Star Omega fluorescent microplate reader. Pelargonidin was tested at different concentration (1, 10, 20 and 50 μM), to evaluate their ability to inhibit the binding of Spike protein (5 nM) to immobilized ACE2, by using the ACE2:SARS-CoV-2 Spike Inhibitor Screening assay Kit. Results are expressed as mean ± SEM (n = 5); * p < 0.05. To confirm the validity of the assay used in this study, we tested plasma samples of post COVID-19 patients as a control. (E) Virus growth in Vero 6E cells analyzed by plaque assay. Pelargonidin was tested at concentration of 20, 50 and 100 μM. To confirm the validity of the assay used in this study, we tested Remdesivir.