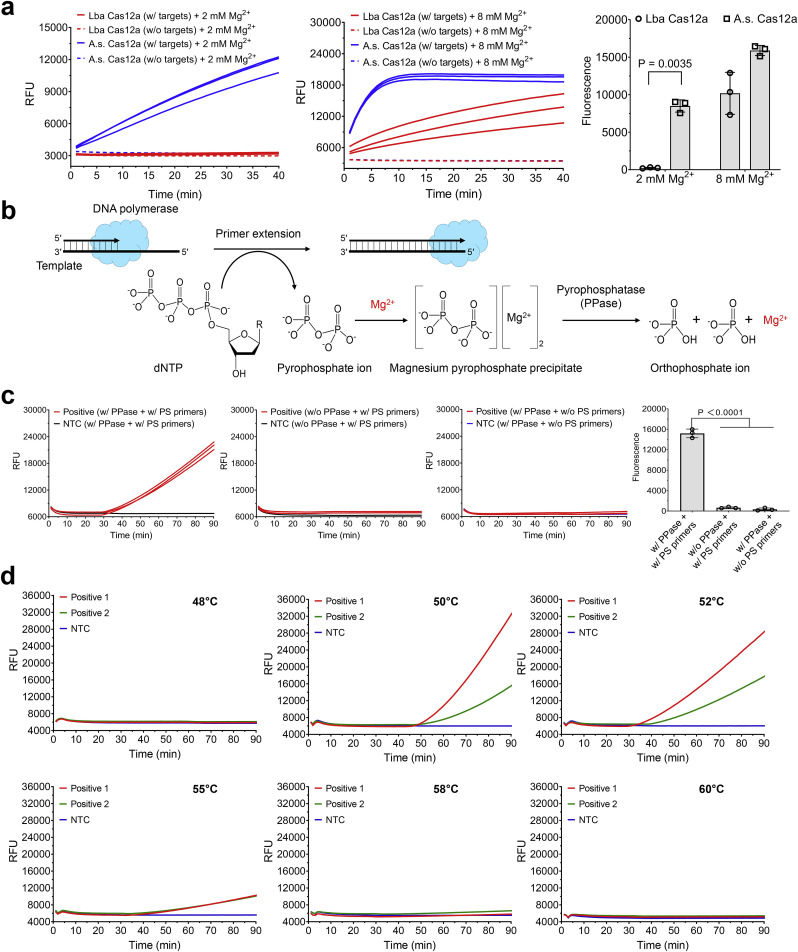
Fig. 2.
Optimization of one-pot WS-CRISPR assay. (a) Comparison of two different Cas12a nucleases for CRISPR-based fluorescence detection at different Mg2+ concentration. Lba Cas12a, EnGen Lba Cas12a from Lachnospiraceae bacterium ND2006 (New England Biolabs). A.s. Cas12a, Alt-R Cas12a (Cpf1) Ultra nuclease from Recombinant Acidaminococcus sp. BV3L6 (Integrated DNA Technologies). The used targets are 1 μM of synthetic SARS-CoV-2 N DNA fragments. Three replicates were run (n = 3). (b) The chemical reaction process of the generation and degradation of the magnesium pyrophosphate precipitate due to the existence of pyrophosphatase during primer extension or DNA polymerization. (c) Real-time fluorescence detection and endpoint fluorescence comparison of one-pot WS-CRISPR assay with PPase and/or PS primers at 52 °C. PS primers specifically denote two phosphorothioated inner primers of FI and RI and the rest of primers are non-phosphorothioated. “w/o PS primers” means reactions with the non-phosphorothioated inner primers. Positive, the reaction with 5 × 104 copies/μl SARS-CoV-2 RNA. Three replicates were run (n = 3). (d) Effect of reaction temperature on one-pot WS-CRISPR assay. Positive 1 and 2, the reactions with 3 × 106 and 5 × 104 copies/μl SARS-CoV-2 RNA, respectively. Three independent assays were conducted with the similar results. NTC, non-template control. Error bars represent the means ± standard deviation (s.d.) from replicates. The statistical significance was analyzed using unpaired two-tailed t-test.