Figure 3.
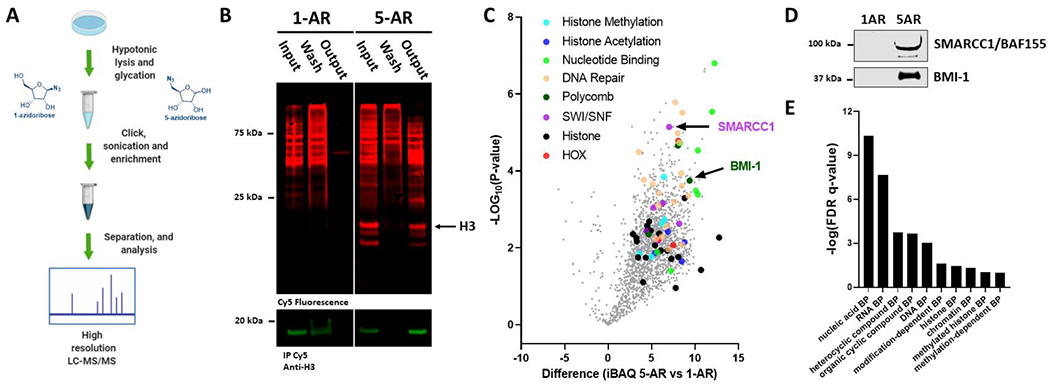
Azidoribose probes can be used to selectively enrich ribose glycation adducts from 293T cells for further analyses. (A) Schematic depicting treatment of isolated nuclei with 5-AR and 1-AR, DBCO Cy5 click labeling, enrichment, digestion, and label-free quantitative proteomics (LFQ) analysis. (B) In-blot fluorescence and western blot showing background labeling and selective enrichment of glycated proteins from AR-treated isolated nuclei. (C) Volcano plot depicting differences in iBAQ values from 5-AR- over 1-AR-enriched sample groups against negative logarithmic (base 10) p-values of the t-test performed from multiple replicates. All identified proteins represented in gray, validated proteins BMI-1 and SMARCC1 labeled, and select protein groups highlighted in separate colors. (D) Western blot of representative output fractions validating the labeling and enrichment of two chromatin remodeling proteomic hits. (E) Binding protein (BP) gene ontology analysis of gene list from set of protein target dataset (p < 0.05 and unique peptides >2).
