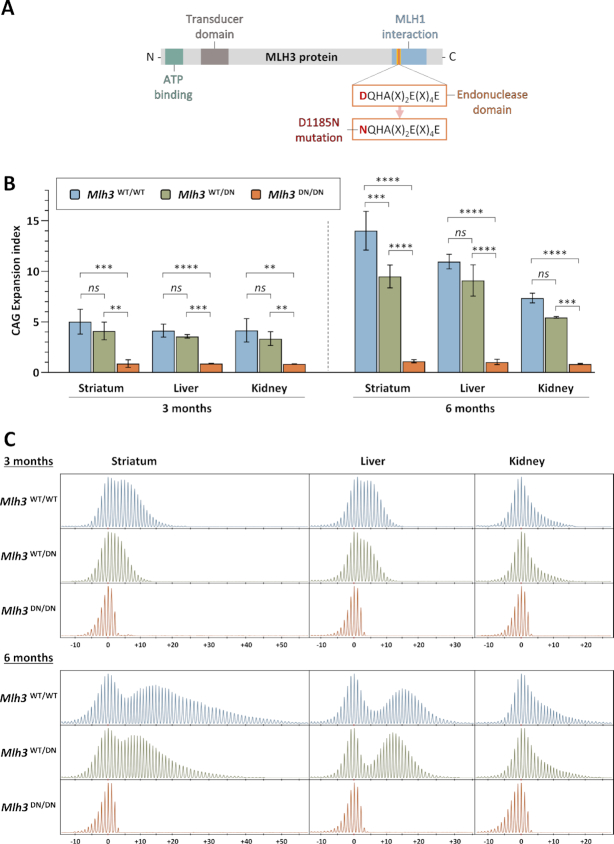
Figure 1.
MLH3 endonuclease domain mutation eliminates HTT CAG expansion in HttQ111 mice. (A) Schematic representation of MLH3 protein and its functional domains, as well as mutation of the endonucleolytic motif in Mlh3DN mice (adapted from Toledo et al. (42)). (B) Quantification of HTT CAG expansions in striatum, liver, and kidney at 3 and 6 months (M) of age in HttQ111 knock-in mice that are either wild type (Mlh3WT/WT, 3M CAG 115–122, 6M CAG 114–121), heterozygous (Mlh3WT/DN, 3M CAG 115–121, 6M CAG 116–121), or homozygous (Mlh3DN/DN, 3M CAG 116–121, 6M CAG 116–121) for the D1185N mutation shows that an intact MLH3 endonuclease domain is required for somatic CAG expansion. Striatum, 3M n = 5–6, 6M n = 3; liver, 3M n = 5, 6M n = 3; kidney, 3M n = 4–5, 6M n = 2–3; error bars represent standard deviation of the mean; two-way ANOVA (with genotype and tissue as variables) with Tukey multiple comparisons post hoc correction. Genotype effect: ns, not significant; **P < 0.01; ***P < 0.001; ****P < 0.0001. (C) Representative GeneMapper traces across different tissues at 3 and 6 months of age in HttQ111 knock-in mice (CAG 121) with different Mlh3 genotypes. 3M: Mlh3WT/WT, CAG 119, Mlh3WT/DN and Mlh3DN/DN, CAG 118. 6M: Mlh3WT/WT, Mlh3WT/DN and Mlh3DN/DN, CAG 121. CAG change from modal allele is represented on the x-axis.