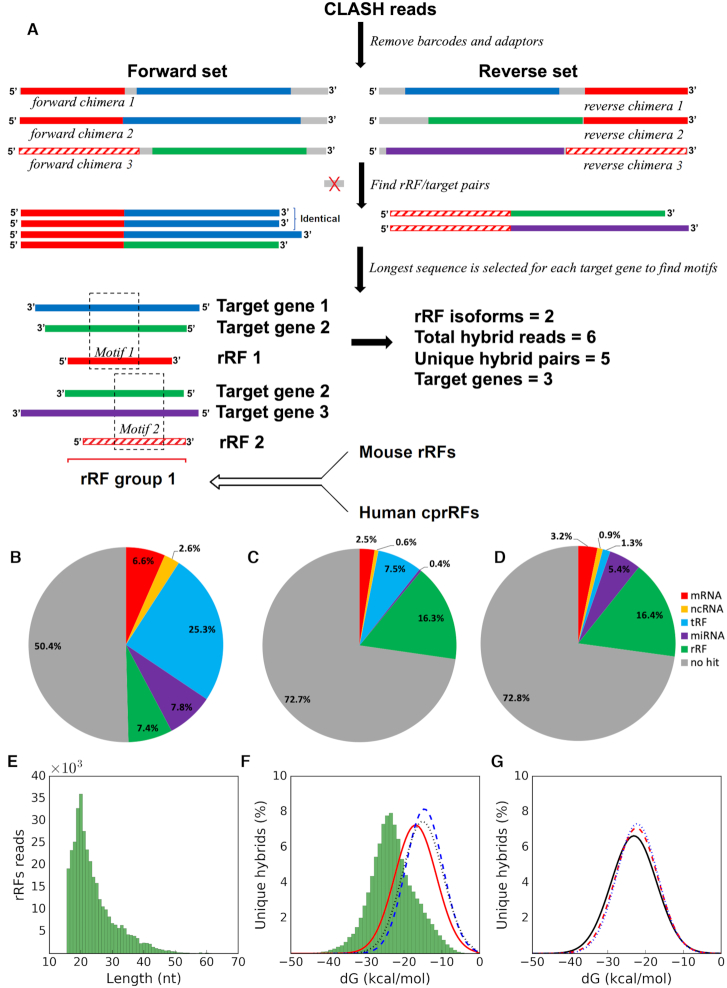
Figure 1.
Analysis and properties of rRFs and other sRNA guides paired with targets in CLASH hybrids. (A) Overview of the meta-analysis of different datasets. Boxes of the same color in CLASH chimeras (top) correspond to the same rRF isoforms or fragments of the same target gene. Gray boxes are nucleotides not matched to rRNA or a target gene. A group (bottom) is produced from overlapping rRFs containing motifs (see section E and Methods) and information from other datasets is integrated with the group to produce a final summary (Supplementary Figure S8). Many more reads (∼106) and unique hybrids (∼105) are typically found and only a few are shown for clarity. Pie-charts show the distributions of various types of targets paired with (B) rRFs, (C) miRNAs and (D) tRFs. Paired target types are color-coded in (B–D), for example, 2.5% of all CLASH targets paired with miRNA guides represent mRNAs (C). Sometimes guide-guide pairs were seen, for example, 0.4% of all CLASH targets paired with miRNA guides represent other miRNAs (C). See Methods for details of the target type assignment. (E) Length distribution of all rRFs identified from CLASH chimeric reads. (F) The MFE distributions of shuffled rRF-target pairs (red solid line) as well as miRNAs (blue dashed line) or tRFs (black dotted line) artificially paired with rRF targets represent randomized controls. The much stronger interactions of specific rRF-target pairs (non-redundant) that we derived from CLASH chimeras are shown as the green histogram. (G) The MFE distributions of rRF-mRNA pairs (black solid line), rRF-tRF pairs (red dashed line) and rRF-miRNA pairs (blue dotted line).