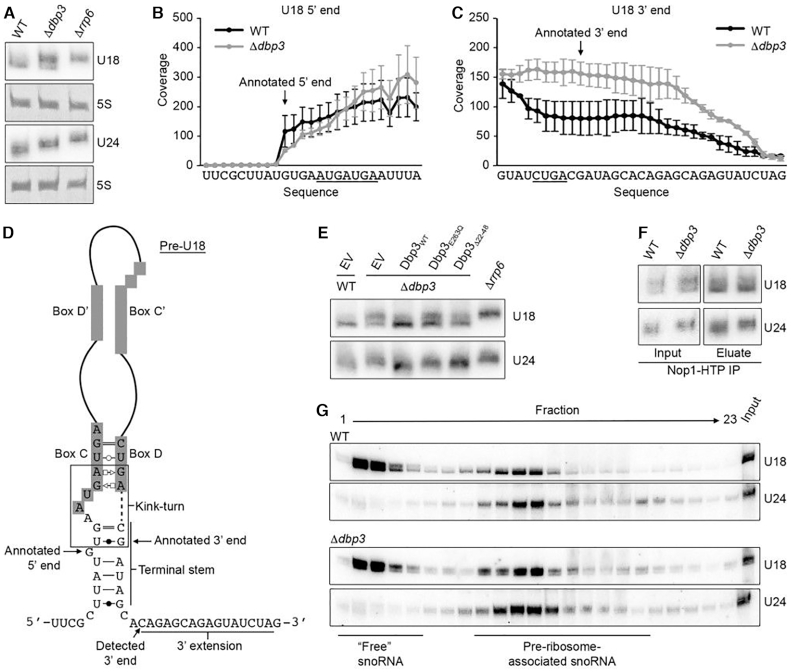
Figure 4.
Maturation of the intron-encoded U18 and U24 snoRNAs requires Dbp3. (A) Total RNA from wild type yeast (WT), and Δdbp3 and Δrrp6 strains was separated by denaturing PAGE and analysed by northern blotting using probes hybridising to the U18 and U24 snoRNAs. (B and C) RMS sequencing reads derived from wild type yeast or the Δdbp3 strain were mapped to the annotated U18 sequence ±50 nt, and after normalization for expression level, the numbers of reads mapping to each nucleotide were determined. Profiles for the 5′ end (B) and 3′ end (C) are shown. Nucleotides of box C (B) and box D (C) are underlined and the annotated 5′ and 3′ ends (61) are indicated. (D) Schematic view of the secondary structure of the pre-U18 snoRNA with key features indicated. Basepairing is shown according to (62). (E) Total RNA from wild type yeast (WT) or a strain lacking Dbp3 (Δdbp3) carrying an empty pRS415 vector (EV) or plasmids for the expression of Dbp3WT, Dbp3E263Q or Dbp3Δ22–48 from the endogenous DBP3 promoter was analysed as in (A). (F) Extracts from yeast expressing Nop1-His-Tev protease cleavage site-ProtA (HTP) in the wild type or Δdbp3 backgrounds were used for pulldown assays on IgG sepharose. Co-purified RNAs were analysed as in (A). (G) Whole cell extracts prepared from wild type yeast or the Δdbp3 strain were separated by sucrose density gradient centrifugation. RNA from individual fractions was separated by denaturing PAGE and analysed by northern blotting using probes to the snoRNAs indicated to the right. The fractions containing ‘free’ snoRNAs and pre-ribosome-associated snoRNAs are indicated.