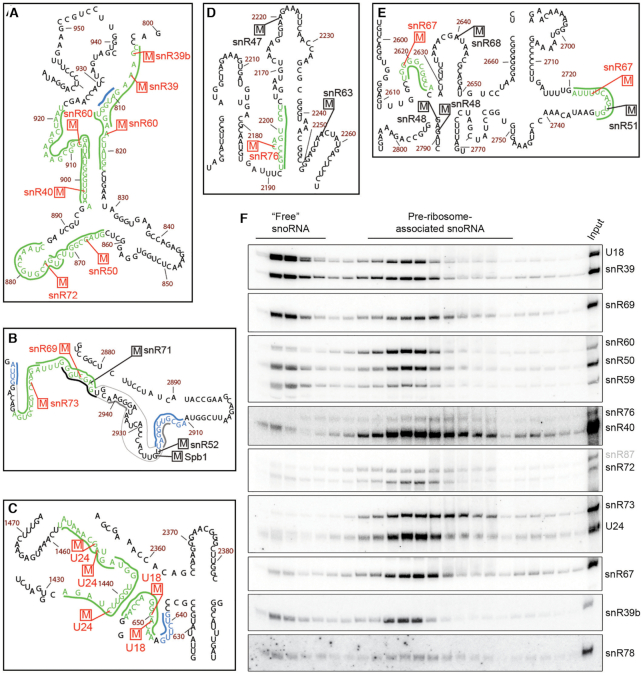
Figure 6.
Many snoRNAs that accumulate on pre-ribosomes in the absence of Dbp3 and guide Dbp3-dependent rRNA 2′-O-methylations have overlapping pre-rRNA basepairing sites. (A–E) 2′-O-methylations reduced in Δdbp3 and the snoRNAs that guide them are highlighted on the secondary structure of the 25S rRNA (55) in red and the pre-rRNA nucleotides involved in basepairing interactions with the guiding snoRNAs are indicated in green. The basepairing of relevant snoRNAs guiding non-Dbp3-dependent rRNA 2′-O-methylations is indicated by thick black lines. Extra basepairing of snoRNAs guiding Dbp3-dependent 2′-O-methylations is shown in blue and that of non-Dbp3 dependent snoRNAs is in grey. Magnified views of selected areas of interest from domain I (A), domain V (B), domain 0 (C), domain IV (D) and domain V (E) are shown. (F) Whole cell extracts prepared from wild type yeast were separated by sucrose density gradient centrifugation. RNA from individual fractions was separated by denaturing PAGE and analysed by northern blotting using probes to the snoRNAs indicated to the right. The fractions containing ‘free’ snoRNAs and pre-ribosome-associated snoRNAs are indicated.