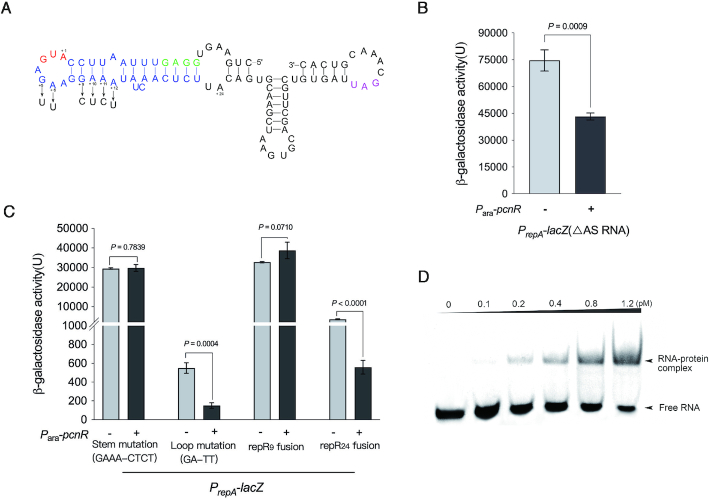
Figure 5.
PcnR regulates the expression of RepA by binding to the repR mRNA. (A) Predicted RNA secondary structure of the repR mRNA. This structure was predicted by RNAfold web server. The Shine-Dalgarno (SD) sequence of repR is shown in green and the stem-loop structure containing the start codon of repR is highlighted in blue. The start codon of repR is shown in red, and A is numbered ‘+ 1’. The stop codon of repR is highlighted in purple. The mutational changes introduced for the present study are shown by arrows. The predicted secondary structure of the leader region of repA mRNA was shown in Supplementary Figure S8B. (B) The inhibition of PcnR on RepA expression is independent of antisense RNA. Activity of PrepA with ΔAS RNA mutation was monitored from the lacZ fusions, and expression of pcnR is controlled by Para with (+) or without (−) arabinose. Error bars represent the SD and P-values were calculated by two-tailed t-tests. (C) The inhibition of PcnR on RepA expression is dependent on the RNA secondary structure of repR mRNA. The stem structure of repR mRNA is predicted to be eliminated by GAAA-CTCT mutation and is denoted by stem mutation (GAAA-CTCT). The GA-TT mutation was located in the loop structure of repR mRNA and is denoted by loop mutation (GA-TT). The repR9 fusion and repR24 fusion represent the lacZ were fused with repR at + 9 and + 24 positions, respectively. Activity of PrepA with these mutations was monitored from the lacZ fusions. Expression of pcnR is controlled by Para with (+) or without (−) arabinose. Error bars represent the SD and P-values were calculated by two-tailed t-tests. (D) The first stem-loop structure of repR mRNA interacts with PcnR protein in vitro. 6  His-tagged PcnR was expressed in E. coli BL21(DE3) from pET28b-pcnR and purified by Ni-NTA affinity chromatography. Purified His-tag-PcnR was cut by thrombin to remove the His-tag and then purified PcnR was used for RNA-EMSA.
His-tagged PcnR was expressed in E. coli BL21(DE3) from pET28b-pcnR and purified by Ni-NTA affinity chromatography. Purified His-tag-PcnR was cut by thrombin to remove the His-tag and then purified PcnR was used for RNA-EMSA.