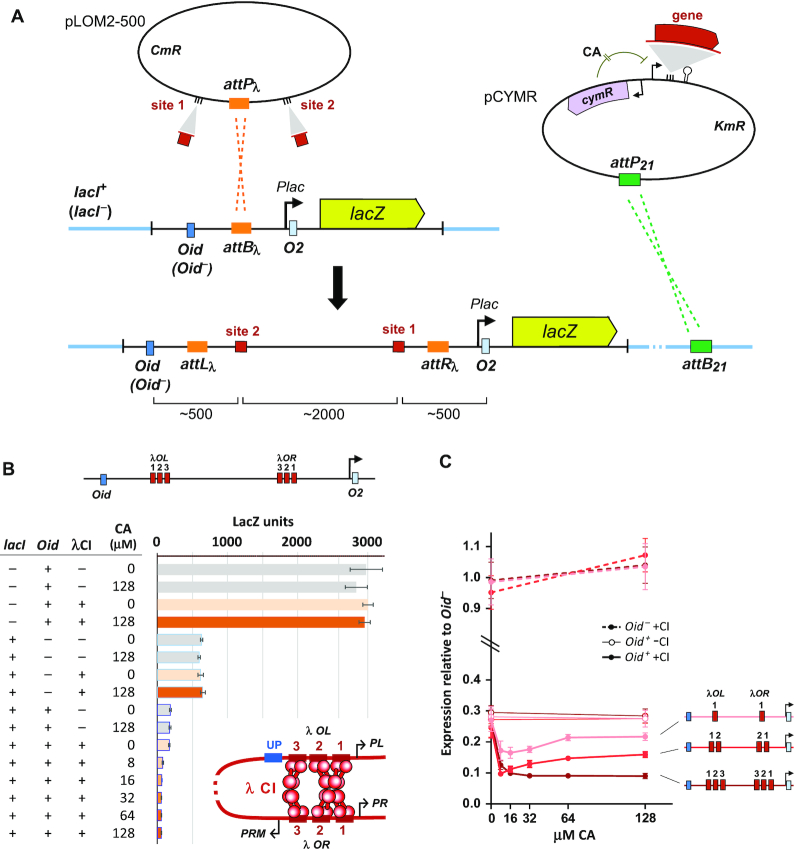
Figure 2.
The loopometer system. (A) The basic procedure for the construction of the looping reporter strains. Protein-binding sites are cloned into the pLOM2–500 plasmid and the resulting plasmid is inserted by λ integrase into a reporter landing pad in the Escherichia coli chromosome, causing the two sites to be nested inside a pair of lac operators, Oid and O2, which control the PlacUV5 promoter for a lacZ.O2– reporter gene. LacI is supplied by a PlacI.lacI+ gene inserted elsewhere in the chromosome. The pLOM2–500 plasmid is also integrated into control strains lacking lacI+ or Oid. The gene for the candidate protein is cloned into the pCYMR expression vector, under the control of the CymR repressor that can be inactivated by cumic acid (CA). This expression vector or the empty control is inserted by φ21 integrase-mediated recombination into the chromosome of the three strains carrying pLOM2–500. (B) LacZ assay results for λ CI and λ OL123-OR123. DNA looping is indicated by a LacI-dependent, Oid-dependent, candidate protein-dependent decrease in units. Error bars are 95% confidence limits (Student's t), n= 6. The insert shows DNA looping by a CI octamer and tetramer at the native OL and OR sites. (C) Graphic display of results for λ CI and OL123-OR123, OL12-OR12, OL1-OR1 reporters (lacI+ only). LacZ units within each set are normalized to the average of the four Oid– values (±CI, ±CA).