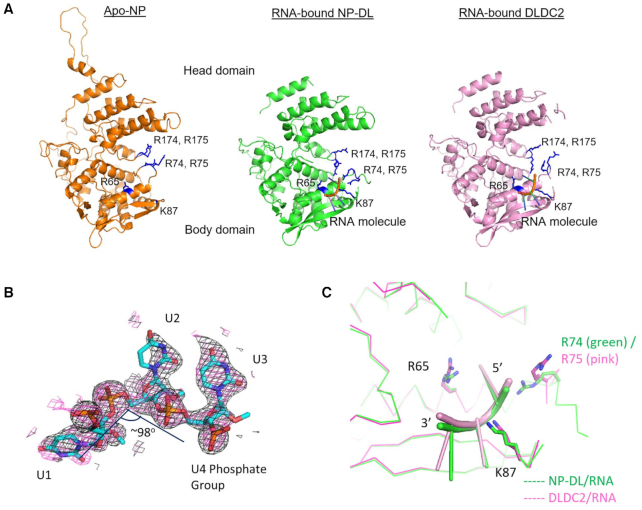
Figure 1.
(A) Apo-NP (PDB 2Q06) and the obligate monomeric NP-DL and DLDC2 shown in the same orientation. Three RNA nucleotides are modeled onto the body domain near the 74–88 loop. The proteins share highly similar tertiary folds demonstrating neither RNA binding nor tail-loop deletion distorts the overall structure. G1 residues (R74, R75, R174, R175), R65 and K87 are marked in blue in the structures. (B) 2Fo – Fc omit maps were generated for the nucleotides modeled on NP-DL (black: simple omit map; pink: simulated-annealing omit map) and contoured at 1σ as shown. (C) The RNA moieties on NP-DL and DLDC2 are situated in a cavity defined by R65, R74/75 and K87. The conformations of two RNA moieties do not deviate significantly. Protein cores are shown as ribbons (NP-DL: green; DLDC2: magenta). RNA is shown as cartoon in green (NP-DL/RNA) and pink (DLDC2/RNA).