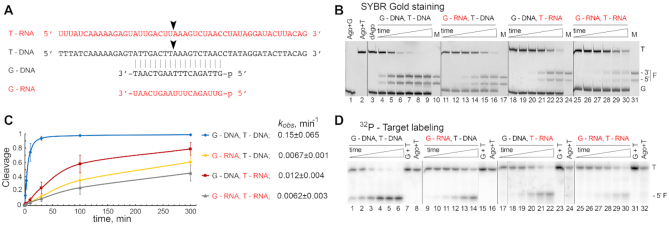
Figure 2.
Guide and target specificity of KmAgo in vitro. (A) Scheme of the guide and target nucleic acids (DNA or RNA); the cleavage position is indicated with an arrow. (B) Analysis of nucleic acid cleavage by KmAgo. KmAgo pre-loaded with guide DNA or RNA was incubated with single-stranded DNA or RNA targets for indicated time intervals, nucleic acids were separated by denaturing PAGE and visualized by SYBR Gold staining. Positions of the targets (T), guides (G) and the 5′- and 3′-cleavage fragments (F) are indicated. The lengths of the synthetic RNA and DNA marker (M) oligonucleotides correspond to the expected cleavage site between the 10th and 11th guide positions. (C) and (D) Kinetics of nucleic acid cleavage by KmAgo measured with radiolabeled target DNA or RNA. The data were fitted to a single-exponential equation. For each guide-target pair, the resulting kobs value is shown (means and standard deviations from three independent measurements). All experiments were performed at 37°C. Representative gels from three independent measurements are shown.