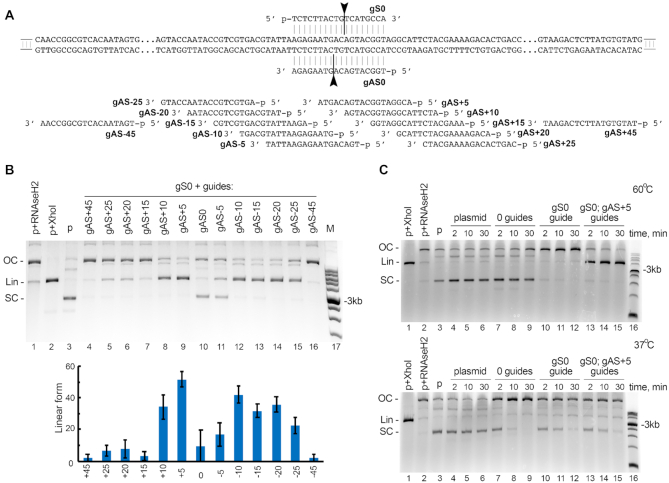
Figure 5.
Cleavage of plasmid DNA by KmAgo. (A) Scheme of the plasmid target used in the cleavage assay (pJET1.2 containing insert 1 shown in Supplementary Table S1). Positions of guide DNAs corresponding to the sense (gS) and antisense (gAS) strands of the target are shown along the plasmid sequence. (B) Plasmid linearization with pairs of guide DNAs with different lengths between the cleavage sites in the two DNA strands. The reactions were performed for 2 min at 60°C. The cleavage efficiency is shown as a fraction of linear DNA form relative to the sum of supercoiled, relaxed and linear DNA (means and standard deviations from three measurements). (C) The target plasmid was incubated in the absence of KmAgo (lanes 4–6), with empty KmAgo (lanes 7–9), or with KmAgo loaded with one or two guide DNAs (gS0 and gAS+5; lanes 10–15) at 60°C (top) or 37°C (bottom) for indicated time intervals. Control samples containing linear (Lin) plasmid obtained by treatment with XhoI (lane 1), relaxed open circle (OC) plasmid obtained by treatment with RNase H2 (lane 2), and supercoiled (SC) plasmid (lane 3) were incubated in the absence of KmAgo. Representative gels from three independent experiments are shown.