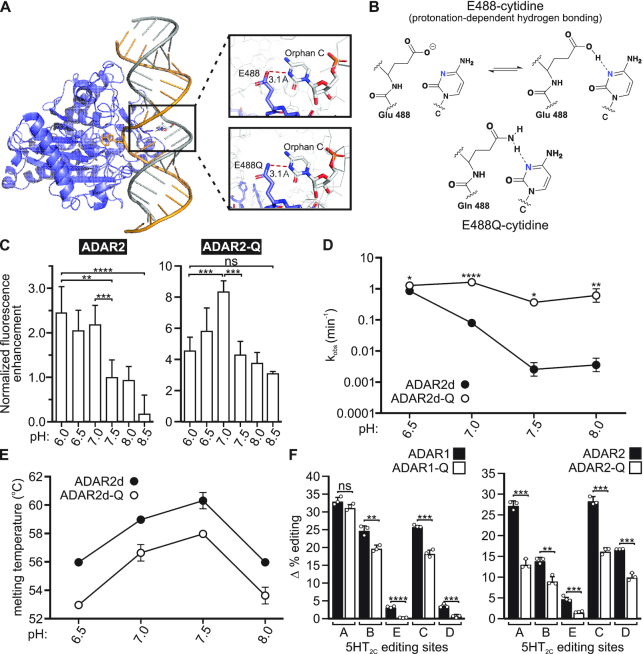
Figure 6.
Effects of a glutamate-to-glutamine substitution on the pH-dependence of ADAR base-flipping and editing. (A) The crystal structure of ADAR2d bound to dsRNA (PDB ID: 5HP3 & 5ED1) shows the base-flipped conformation stabilized by contacts between residue 488 and the orphan base. (B) An illustration of the hydrogen bonding contact between ADAR2 and the orphan base showing protonation-dependent hydrogen bonding for wild-type ADAR2. (C) Normalized fluorescence enhancement from a dsRNA substrate containing 2-aminopurine in the edited position, corresponding to base-flipping by the ADAR2 enzyme. Plotted values represent the means of three technical replicates ± SD. Statistical significance between groups was determined using one-way ANOVA with Tukey's multiple comparisons test; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; ns, not significant. (D) Rates of in vitro ADAR2d- and ADAR2d-Q mediated Gli1 (+23 site) editing. Plotted values represent the means of three technical replicates (○) ± SD. Statistical significance between groups was determined using the Holm-Sidak t-test for multiple comparisons; *P ≤ 0.05; **P ≤ 0.01; ****P ≤ 0.0001. (E) Quantification of ADAR2d and ADAR2d-Q melting temperatures across the pH range used for in vitro editing experiments. Plotted values represent the means of three technical replicates (○) ± SD. (F) Quantification of the extent of acidification-induced increases in 5HT2C editing mediated by wild-type ADARs or ADAR-Q mutants (Δ % editing = % site-selective editing at pH 6.7 - % site-selective editing at pH 7.4). Plotted values represent the means of three biological replicates (○) ± SD. Statistical significance between groups for each 5HT2C site was determined using the Holm–Sidak t-test for multiple comparisons; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.