Fig. 4.
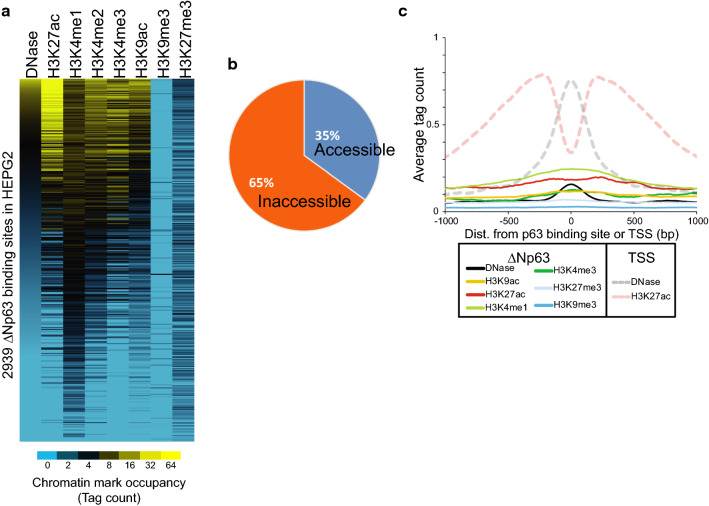
ΔNp63 expression in HepG2 leads to binding at inaccessible and unmodified chromatin regions. a The 2939 ΔNp63-bound sites in HepG2 cells with their histone modifications and DNase measurements. Sites are sorted by DNase signal. Chromatin datasets are from HepG2 in the absence of ΔNp63 from the ENCODE project. b Chromatin accessibility for the 2939 ΔNp63-bound sites in HepG2. Accessibility is defined from the synthesis track of DNase and FAIRE from ENCODE (GSM1002654) [66]. c Average chromatin architecture in HepG2 at inaccessible ΔNp63 ChIP-seq summits compared to transcriptional start sites (TSS). Data are plotted for 2 kb flanking the summit or TSS