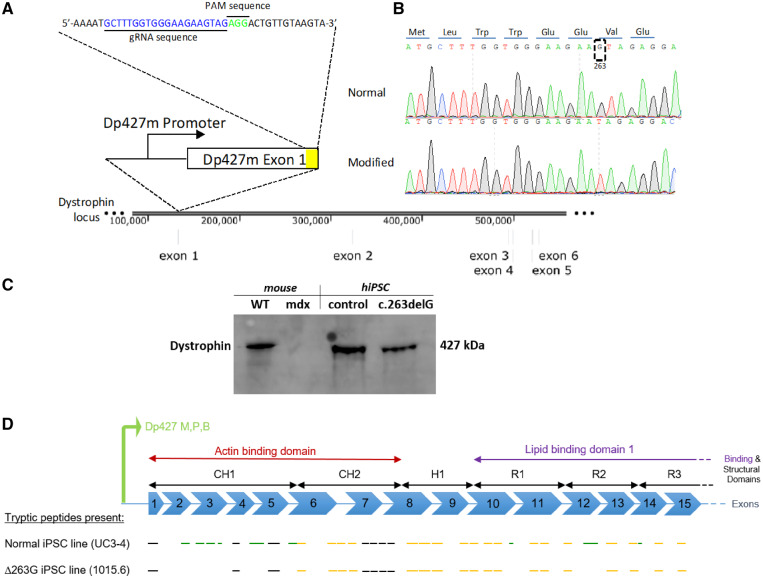
Figure 1.
Creation of a dystrophin mutant human iPSC line with an isogenic background. (A) CRISPR-Cas9 enzyme was used to target the dystrophin exon 1 in a normal iPSC line to create an isogenic dystrophin-modified line. (B) Sangers sequencing trace illustrates the location around the modified region in both normal and modified hiPSC lines. The translated dystrophin N-terminus protein is shown at the top. (C) Cropped panel of antibody targeting C-terminal domain of dystrophin in WT and mdx mouse cardiac tissue and control (UC3-4) and dystrophin mutant (c.263delG) hiPSC lines. Dystrophin bands at 427 kDa were detected in WT, control and, surprisingly, in c.263delG. Uncropped gel reported in Supplementary material online, Figure S1C. (D) Schematic representation of Dp427 promoter for muscle, purkinje and brain to be consistent with Supplementary material online, Figure S2. Presumed mass spectroscopy observable tryptic peptides are shown by black line segments (but were not detected in either cell line), peptides only seen in the normal UC3-4 hiPSC cell line shown in green and those peptides present in both normal and UC-1015.6 hiPSC line (c.263delG) are shown in dark yellow.