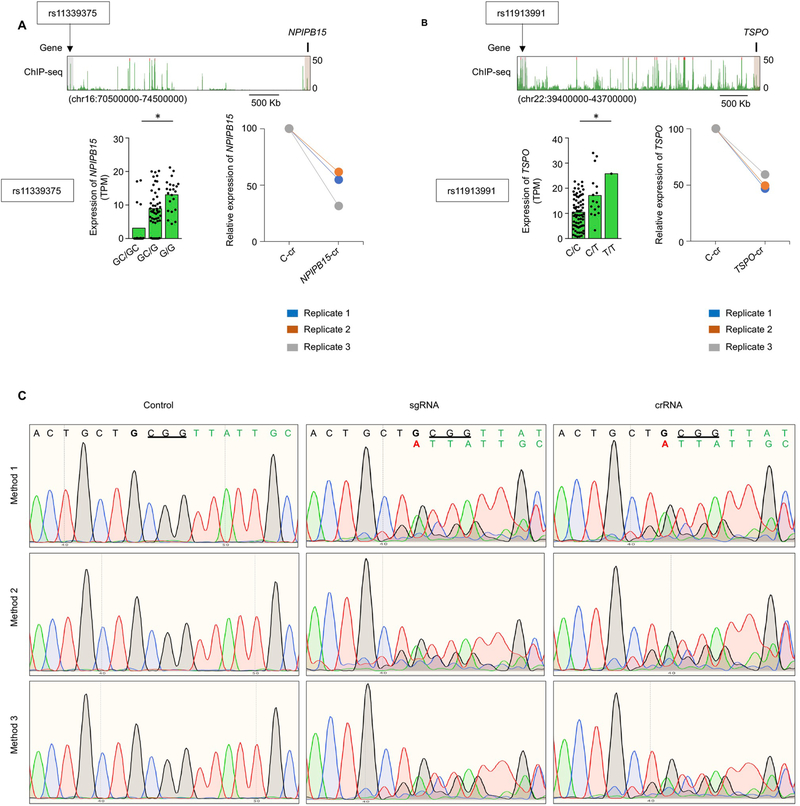
Extended Data Fig. 7 |. Validation of ultra-long pieQtL, Related to Fig. 3.
a, WashU Epigenome browser tracks for the extended NPIPB15 locus and H3K27ac ChIP-seq tracks for the indicated locus. Bottom left panel, mean expression levels (TPM) of NPIPB15, eGenes in naïve B cells from subjects (n=91) categorized based on the genotype at the indicated cis-eQTL; each symbol represents an individual subject; * adj. association P value: 0.007, calculated by Benjamini-Hochberg method. Bottom right panel, real-time PCR quantification of NPIPB15 transcript levels (relative to the housekeeping gene YWHAZ) in GM12878 cells 48 hour after CRISPRi-mediated silencing of indicated enhancer (n=3). b, WashU Epigenome browser tracks for the extended TSPO locus and H3K27ac ChIP-seq tracks for the indicated locus. Bottom left panel, mean expression levels (TPM) of TSPO, eGenes in naïve B cells from subjects (n=86) categorized based on the genotype at the indicated cis-eQTL; each symbol represents an individual subject; * adj. association P: 0.03, calculated by Benjamini-Hochberg method. Bottom right panel, real-time PCR quantification of TSPO transcript levels (relative to the housekeeping gene YWHAZ) in GM12878 cells 48 hours after CRISPRi-mediated silencing of indicated enhancer (n=3). c, CRISPR-mediated homology-directed recombination (HDR) in primary human T cells from a donor homozygous for the rs11130745G/G allele for the FHIT-related 1.2 Mb away ultra-long pieQTL. The sanger sequencing result of CRISPR-mediated genome editing of rs11130745G/G allele to rs11130745A/A allele with a deletion of the adjacent 3 bp PAM sequence by three independent methods using control RNA, sgRNA and crRNA is shown.