Abstract
Background
siRNA brings hope for cancer therapy. However, there are many obstacles for application of siRNA in clinical. Because of the excellent biocompatibility, non-toxicity and non-immunogenicity of bovine serum albumin (BSA), BSA-based nanoparticles have been widely designed as a drug carrier system.
Methods
The optimal formula for BSA NPs preparation was investigated by central composite design response surface methodology (CCD-RSM), BSA-based survivin-siRNA delivery system (BSA NPs/siRNA) was characterized by dynamic light scattering, atomic force microscope, transmission electron microscope and Bradford method. The in vitro anti-tumor effect and mechanism of BSA NPs were investigated by confocal microscopic imaging, MTT assay, RT-qPCR and ELISA analysis. Moreover, the anti-tumor effect, distribution and biosafety of BSA NPs were studied in vivo.
Results
The optimal formula for BSA NPs was settled to be 20 mg/mL for BSA concentration, 9 for pH value, 136% for crosslinking degree and 1.6 mL/min for speed of ethanol addition. BSA NPs/siRNA could remain stable at 4°C for 4 weeks and protect siRNA from degradation by RNase A. Besides, BSA NPs/siRNA could maintain a sustained release of siRNA and promote the uptake of siRNA significantly. The survivin-mRNA level and the survivin-protein level were decreased by 55% ± 1.6% and 54% ± 1.6% separately. The in vivo tumor inhibition results suggested that the tumor inhibition rate of BSA NPs/siRNA-treated group was 54% ± 12% and was similar with that of DOX-treated group (57% ± 9.2%, P > 0.05). The biosafety results confirmed that BSA NPs/siRNA could not induce significant damages to the main organs and blood in vivo.
Conclusion
These results demonstrated that CCD-RSM was an effective tool for preparation analysis, and the BSA NPs/siRNA was a promising system for siRNA-based gene therapy.
Keywords: bovine serum albumin, BSA, CCD-RSM, survivin-siRNA, nanoparticles, RNAi
Introduction
RNA interference (RNAi) has attracted a broad attention since its discovery in the 1990s.1 Oligonucleotides, such as small interfering RNAs (siRNAs), are promising drug modality for a variety of diseases.2 As a 21–23 bp double-stranded RNA molecule, siRNAs could induce a sequence-specific posttranscriptional gene silencing by targeting complementary nucleotide sequence.3 However, there are still many obstacles for application of siRNAs in clinical.4 The large molecular and negative charge will promote the renal clearance, and prevent the cellular transmembrane of siRNAs after administration.5 Moreover, siRNAs are easy to be degraded during blood circulation, and will cause serious immune response in vivo.6 So it has great significance to design a siRNA delivery system with excellent transfection effect and good biocompatibility.7 Survivin gene is a member of the inhibitor of apoptosis protein family, which has the ability to inhibit cell apoptosis and regulate cell division. The expression of Survivin gene has highly tumor specificity, making it an ideal target for tumor gene therapy. According to previous research, inhibition of Survivin gene was an effective method for treatment of various tumors.8–10
Bovine serum albumin (BSA) is an endogenous biomacromolecule, and has been widely used as a material for drug delivery.11,12 Compared to other carrier materials, BSA has many advantages, such as biodegradability, non-toxicity, and non-immunogenicity.13,14 Moreover, BSA has a dual targeting ability to tumor tissues. As reported, BSA could bind with albumin receptor gp60 and the cysteine rich acidic secretory protein (SPARC), both of which are highly expressed in tumor tissues.15 Considering the remarkable characters of BSA, it has broadly applied for delivering of chemical drugs, gene drugs, antibodies and so on.16 It is reported that BSA nanoparticles could encapsulate genes by the abundant positive amino acids in BSA.17 Although BSA has been investigated as a drug delivery carrier in a lot of researches, few study was carried out to optimize the preparation condition of BSA nanoparticles.
Central composite design response surface methodology (CCD-RSM) is an effective method for multi-factor experimental analysis, and is frequently applied for prescription optimization in the field of pharmacy.18 Compared with other factor analysis methods, CCD-RSM is more accurate and convenient.19 So it is usually adopted to investigate the influence of multiple factors on the results of pharmaceutical preparation.20–22
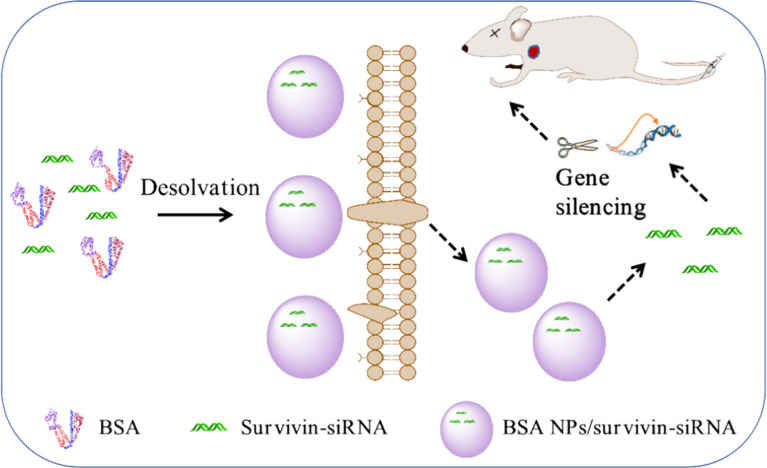
In this research, CCD-RSM was carried out to optimal the prescription of BSA NPs, and the optimal formula was applied to prepare survivin-siRNA loaded BSA NPs (BSA NPs/siRNA). As shown in Figure 1, BSA NPs could not only protect survivin-siRNA from degradation but also enhance the gene silencing effect of survivin-siRNA by cellular-uptake promoting effect and targeting effect of BSA. As an inhibitor of apoptosis protein family highly expressed in tumor, silencing of survivin gene would exert an efficient anti-tumor effect. It is highly expected that the BSA NPs are a promising carrier for targeted gene delivery, and have a huge application potential for RNAi therapy.
Figure 1.
The mechanism of BSA NPs/siRNA preparation and anti-tumor effect. BSA NPs/siRNA were prepared by desolvation-crosslinking method. BSA NPs/siRNA could target survivin-siRNA into tumor tissues, and promote the cellular uptake of survivin-siRNA. The gene silencing effect of survivin-siRNA was significantly enhanced after loaded in BSA NPs.
Materials and Methods
Materials
Bovine Serum Albumin, Glutaraldehyde solution (50 wt % in H2O) and Proteinase K were purchased from Sigma-Aldrich (St. Louis, MO, USA). Survivin-siRNA, Cy3-survivin-siRNA and Cy5-survivin-siRNA were purchased from GL Biochem Ltd. (Shanghai, China). LipoTM2000 (Lipo), TRIzol and Hoechst 33342 were purchased from Invitrogen (Carlsbad, CA, USA). TaqMan® Gene Expression Master Mix, High Capacity RNA-to-cDNA Kit, High Capacity cDNA Reverse Transcription Kit, Human Survivin Quantikine ELISA Kit and TaqMan® Gene Expression Assays were obtained from Thermo Fisher Scientific (Waltham, MA, USA). MCF-7 cell line was purchased from the Cell Bank of Chinese Academy of Medical Sciences. MCF-7 cells were cultured in RPMI-1640 medium (Hyclone Laboratories Inc., Logan, USA) with 10% Fetal bovine serum (Gibco, Australia). The cells were detached with 0.25% Trypsin (Hyclone, USA). All reagents were analytical grade and deionized water was used in all experiments.
The sequences of homo-survivin-siRNA are 5ʹ-CACCGCAUCUCUCUACAUUC ATT-3ʹ (sense) and 5ʹ-UGAAUGUAGAGAGAUGCGGUGTT-3ʹ (anti-sense). The sequences of mus-survivin-siRNA are 5ʹ-CCGAGAACGAGCCUGAUUUTT-3ʹ (sense) and 5ʹ-AAAUCAGGCUCGUUCUCGGTT-3ʹ (anti-sense). The sequences of NC (negative control) are 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ (sense),5ʹ-ACGUGACACGUUCGGAGAATT −3ʹ (anti-sense).
Preparation and Formulation Optimization of BSA NPs
Bovine serum albumin nanoparticles (BSA NPs) were prepared by desolvation-crosslinking method.23–25 Firstly, bovine serum albumin (BSA, 50 mg) was dissolved in NaCl (2 mL, 10 mM) solution, and stirred for 15 minutes at room temperature. The pH of BSA solution was adjusted to 8.0, and ethanol was added dropwise (1 mL/min) until the solution appearing light blue opalescent. The nanoparticles solution was stirred for 3 h at room temperature, and then glutaraldehyde aqueous solution (20 μL, 8%) was added to obtain BSA NPs. After stirring for 24 h at room temperature, the nanoparticles suspension was purified by centrifugation (12000 ×g, 30 min) for three times.
Six basic factors related to the preparation of BSA NPs were investigated by variable-controlling approach. That is BSA concentration, pH value, glutaraldehyde concentration, volume of ethanol, speed of ethanol addition and stirring rate. The hydrodynamic diameter and polydispersity index (PDI) were measured to evaluate the effects of the factors.
Then four factors that significantly influenced the preparation of BSA NPs were selected. That is BSA concentration, pH value, glutaraldehyde concentration and speed of ethanol addition. For each factor, five levels were designed. And total of 30 groups were carried out to optimize the preparation of BSA NPs. Hydrodynamic diameter, PDI and zeta potential were applied as evaluation indexes. The results were analyzed by central composite design (CCD) response surface methodology, and calculated by Design Expert.26
Preparation of BSA NPs/siRNA
The optimal formulation obtained above was used to prepare BSA NPs/siRNA. Twenty-milligram BSA was dissolved in NaCl solution (1 mL, 10 mM), and was stirred for 15 minutes at room temperature. Survivin-siRNA (1.2 nmoL) was added to BSA solution, and stirred for 1 h at room temperature. The pH of the solution was adjusted to 9.0, and ethanol was added dropwise (1 mL/min) until the solution appearing light blue opalescent. The nanoparticles solution was stirred for 3 h at room temperature, and then glutaraldehyde aqueous solution (16 μL, 8%) was added to obtain BSA NPs. After stirring for 24 h at room temperature, the nanoparticles suspension was purified by centrifugation (12000 ×g, 30 min) for three times. The precipitate was resuspended in deionized water solution.
Characterization of BSA NPs and BSA NPs/siRNA
The hydrodynamic diameter, PDI and zeta potential of BSA NPs/siRNA were measured by dynamic light scattering (DLS, Nano ZS90, Malvern Panalytical, UK). Surface morphology of the nanoparticles was observed by atomic force microscope (AFM, Veeco Instruments Inc., Plainview, NY, USA) and transmission electron microscope (TEM, JEOL, Tokyo, Japan).
Nanoparticles Yield and Survivin-siRNA Loading
The yield of BSA NPs was measured by Bradford method.27 After preparation by desolvation-crosslinking method, BSA NPs solution was centrifuged by 12000 ×g, and the supernatant was obtained. The total protein for preparation of BSA NPs (Total protein) and the protein in supernatant (Protein in supernatant) were tested by coomassie brilliant blue (CBB). And the nanoparticle yield was calculated as follows:
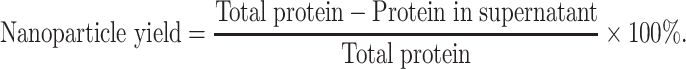 |
The amount of survivin-siRNA loaded in BSA NPs/siRNA was determined by electrophoresis (CZE, MDQ, Beckman).28 BSA NPs/siRNA were centrifuged by 12000 ×g, and the supernatant was obtained. The amount of survivin-siRNA in the supernatant was measured as the following experimental parameters: the fused-silica capillary with effective length of 40 cm and inner diameter of 50 μm, 30 kV for potential, temperature 30°C, 12.5 mM of borate buffer, pH 9.5, 260 nm for the wave length of the diode-array detector. And the survivin-siRNA loading capacity of BSA NPs/siRNA was calculated as follows:
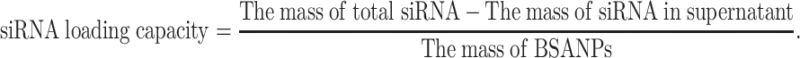 |
Release Profile of BSA NPs/siRNA
Release profile of siRNA from BSA NPs/siRNA was measured by dialysis. FAM labelled survivin-siRNA were used. Naked siRNA and BSA NPs/siRNA (500 μL, 100 nM) were dispersed in 500μL PBS solution (pH 7.4), and placed into dialysis membranes (MWCO: 8–14 kD) respectively. The dialysis was conducted in 5mL PBS solution at 37°C. The 5mL PBS dialysis solution was collected and replaced with fresh dialysis solution at 1 h, 2 h, 5 h, 10 h, 24 h, 36 h, 48 h, 60 h, 72 h, 96 h, 120 h, 144 h, respectively. The amount of survivin-siRNA was measured by fluorescence spectrophotometer (F-2500, HITACH, Japan, 492 nm for excitation, 518 nm for emission), and the cumulative release of survivin-siRNA was calculated.
Stability of BSA NPs/siRNA
BSA NPs/siRNA were placed at 4°C for 4 weeks. And the hydrodynamic diameter and PDI were measured once every week to evaluate the storage stability of BSA NPs/siRNA.
The enzymatic stability of BSA NPs/siRNA was evaluated by agarose gel retardation assay. BSA NPs/siRNA and survivin-siRNA were treated with RNAse (10 µg/mL) separately for 0.5, 1 and 2 h at 37°C. And then Proteinase K (100 µg/mL) was applied to release survivin-siRNA from BSA NPs/siRNA. The supernatant was collected for determination. The survivin-siRNA levels in all samples were measured by agarose gel retardation assay.29
Cell Culture
MCF-7 human breast tumor cells were cultured in RPMI 1640 medium supplemented with 10% FBS, penicillin (100 U/mL) and streptomycin (100 μg/mL) at 37°C with 5% CO2. The cells were subcultured 2–3 times a week till they reached 80% confluence.
Cellular Uptake
Cy3-labelled survivin-siRNA (Cy3-siRNA) was applied to observe the location of siRNA in cells. MCF-7 cells were seeded in 20 mm confocal dishes at a density of 2×105 cells/mL. Then, the cells were treated with Cy3-siRNA (100 nM), BSA NPs/Cy3-siRNA (contain 100 nM Cy3-siRNA) and Lipo/Cy3-siRNA (contain 100 nM Cy3-siRNA) separately. After transfection for 4 h, the medium in each dishes was removed. And then Hoechst 33342 (4 μg/mL) was added. After incubation for another 15 minutes, the staining solution was discarded and the cells were washed with PBS solution for 3 times. The cellular uptake results were observed by a confocal laser scanning microscopy (CLSM, TCSSP5, Leica, Wetzlar, Germany).30
MTT Assay
MTT assay was used to evaluate the cytotoxicity of BSA NPs and the proliferation inhibition effect of BSA NPs/siRNA.31 MCF-7 cells (5×103 cells/well) were seeded in a 96-well plate and incubated for 24 h. For cytotoxicity assay of BSA NPs, the cells were treated with 100 μL BSA NPs with different concentrations (0.1, 0.2, 0.4, 0.6, 0.8, 1.0, 2.0, 4.0 mg/mL). For anti-proliferation assay of BSA NPs/siRNA, the cells were treated respectively with siRNA, Lipo/NC, BSA NPs/NC, Lipo/siRNA and BSA NPs/siRNA (contain 30, 60, 90 and 120 nM siRNA). All cells were incubated for 48 h at 37°C with 5% CO2. Then, 25 μL MTT solution (5 mg/mL) was added into each well, and cells were incubated for another 4 h. Then the medium was removed, and DMSO solution (150 μL) was added. The absorbance was measured at 570 nm by a microplate reader (EnSpire, PerkinElmer Inc., Waltham, MA, USA).
RT-qPCR
The expression of survivin-mRNA was measured by RT-qPCR.32 MCF-7 cells were seed in six-well dishes at the density of 2×105 cells/well for 12h. After washing 2 times with PBS, the cells were treated with 100 nM of siRNA, Lipo/NC, BSA NPs/NC, Lipo/siRNA and BSA NPs/siRNA separately for 6 h. Then the solution in dishes was replaced by completed medium, and the cells were incubated for another 42 h. TRIzol reagent was applied to exact the total RNA. GeneAmp® PCR System 9700 (Applied Biosystems, USA) and Real-Time PCR System 7500 (Applied Biosystems, USA) were separately used to reverse RNA to cDNA and amplify the expression of cDNA. NanoDrop Spectrophotometer (ND-1000, Thermo Scientific, USA) was applied for the measure of concentration of RNA and cDNA. The value relative quantity of survivin-mRNA expression was calculated with average threshold cycle (Ct) by the delta-delta Ct (2−ΔΔCt) method.
ELISA
The expression of survivin protein was measured by ELISA.33 MCF-7 cells were seed in six-well dishes at the density of 2×105 cells/well for 12h. After washing 2 times with PBS, the cells were treated with 100 nM of siRNA, Lipo/NC, BSA NPs/NC, Lipo/siRNA and BSA NPs/siRNA separately for 6 h. Then the solution in dishes was replaced by completed medium, and the cells were incubated for another 42 h. The total protein was obtained by a protein extraction solution (RIPA/PMSF/Cocktail=100/1/1). The concentration of cell protein was measured by a BCA Protein Assay kit, and the amount of survivin-protein was measured by a Human Survivin ELISA kit according to the instructions. Plates were read at 450 nm by a microplate reader to measure the optical density (OD) values.
Tumor Inhibition of BSA NPs/siRNA in vivo
Female BALB/c nude mice (four weeks, 13–15 g) were obtained from Animal Department of Capital Medical University and raised at SPF environment (temperature: 22–25°C, humidity: 50 ± 2.0%). All of the animal experiments were approved by the Institutional Animal Ethics Committee of Capital Medical University, and the welfare of the laboratory animals was “The Laboratory Animal-Guideline for Ethical Review of Animal Welfare issued by the National Standard GB/T 35892–2018 of the People’s Republic of China”.
The mice were intraperitoneally injected with 0.2 mL estrogen (0.75 mg/mL) every three days. After two weeks, MCF-7 cells (1×107 cells/mouse) were subcutaneously injected into the axilla of mice to establish breast cancer xenografts. When the tumor volume (volume (mm3) = length × width2/2) reached approximately 150 mm3, all the nude mice were randomly divided into four groups with 6 mice in each group. Each group was intravenously injected with normal saline (Control), 0.3 mg/kg survivin-siRNA (siRNA), BSA NPs/siRNA (contain 0.3 mg/kg survivin-siRNA) and 1.2 mg/kg DOX respectively every other day for 5 times. The tumor volume of the nude mice were recorded every other day. On the day after the last injection, all nude mice were weighted and sacrificed. The tumor of each mice was collected and photographed. And the main organs (heart, liver, spleen, kidney and brain) were also harvested to study organ toxicity of BSA NPs/siRNA.34
Distribution of BSA NPs/siRNA in vivo
Breast cancer xenografts BALB/c nude mice were randomly divided into three groups. Each group was intravenously injected with normal saline (Control), 0.3 mg/kg Cy5-survivin-siRNA (siRNA) and BSA NPs/siRNA (contain 0.3 mg/kg Cy5-survivin-siRNA) respectively. The fluorescence imaging of the nude mice were photographed at 0.5, 4 and 8 h by an in vivo fluorescence imaging system (IVIS Spectrum, PerkinElmer Inc., Waltham, MA, USA).
Blood Biochemistry Analysis
BALB/c nude mice (6 weeks old, female, about 15 g) were divided into four groups (n=5): normal saline group (control), naked survivin-siRNA group (siRNA, 0.3 mg/kg), BSA NPs/siRNA group (contain 0.3 mg/kg siRNA), and DOX group (1.2 mg/kg). After drug administration every other day for 5 times, blood samples were taken from each nude mouse. The complete blood panel data was measured by an automatic hematology analyzer (MEK6400, Nihon Kohden, Japan). And the main blood biochemical indicators were detected by an automatic biochemical analyzer (BS-350E, Mindray, China).35
Statistics Analysis
Data was expressed as mean ± SD (standard deviation) based on at least three times experiments. t-test was used for statistical analysis.
Results
Investigation of Prescription Optimization
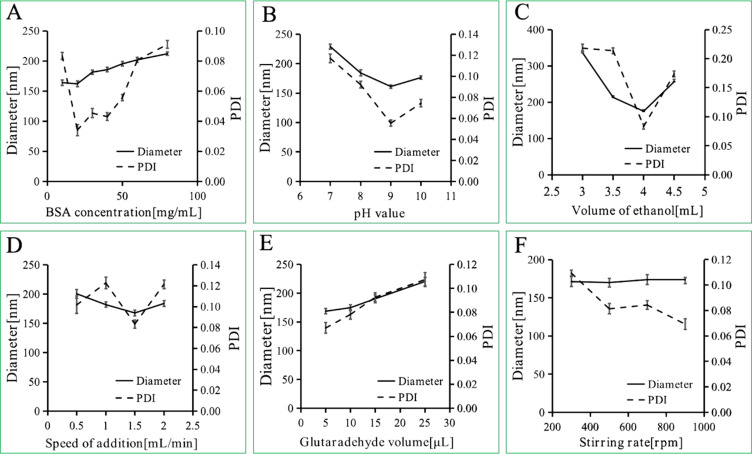
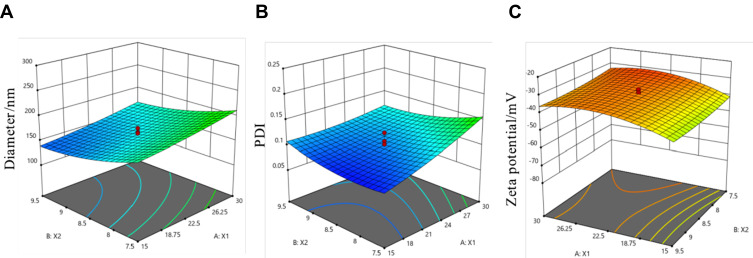
Six factors in preparation of BSA NPs were investigated in this research. As shown in Figure 2, BSA concentration, pH value, ethanol volume, speed of ethanol addition and glutaraldehyde volume were the most significant factors for BSA NPs preparation. Since ethanol volume is related to the other factors, it is not suitable for multi-factor experimental analysis. So the multi-factor experiments were designed based on the other four factors (BSA concentrations, pH values, speed of ethanol addition and glutaraldehyde volume). Five levels of the four factors were designed through CCD-response surface methodology, and total of 30 assays were generated by Design Expert (Table 1). The experimental results suggested that BSA concentration and pH value were the most important factors for BSA NPs preparations. So the response surface plots for hydrodynamic diameter (Figure 3A), polydispersity index (Figure 3B) and zeta potential (Figure 3C) were investigated to study the interaction between BSA concentration and pH value, as well as got the optimal BSA concentration and pH value for BSA NPs preparation. The results suggested that BSA NPs had the best character when BSA concentration was 20 mg/mL and pH value was 9. So, according to the analysis above, the optimal formula for BSA NPs preparation was settled to be 20 mg/mL for BSA concentration, 9 for pH value, 136% for crosslinking degree and 1.6mL/min for speed of ethanol addition.
Figure 2.
Diameter and PDI of BSA NPs prepared by different BSA concentrations (A), pH values (B), ethanol volume (C), speed of ethanol addition (D), speed of stirring rate (E), glutaraldehyde concentrations (F).
Table 1.
The Experiments Designed by CCD-RSM
| Factors | Symbol | Coded Levels | ||||
|---|---|---|---|---|---|---|
| −2 | −1 | 0 | 1 | 2 | ||
| BSA concentration/mg/mL | X1 | 7.5 | 15.0 | 22.5 | 30.0 | 37.5 |
| pH value | X2 | 6.5 | 7.5 | 8.5 | 9.5 | 10.5 |
| Glutaradehyde concentration/% | X3 | 25 | 75 | 130 | 175 | 225 |
| Speed of ethanol addition/mL/min | X4 | 0.5 | 1.0 | 1.5 | 2.0 | 2.5 |
Notes: Four main factors (BSA concentration, pH value, speed of ethanol addition and glutaraldehyde volume) were investigated; five levels were designed for each factor.
Figure 3.
Response surface plots for diameter (A), PDI (B), zeta potential (C), and the plots indicated the interaction between X1 and X2. X1: BSA concentration (mg/mL), X2: pH value.
The optimal formula was applied for the preparation of BSA NPs. And the diameter of BSA NPs prepared was 161.8 ± 5.6 nm, the PDI was 0.084 ± 0.004, and the zeta potential was −34.10 ± 2.5 mV. The measured values had no significant difference with the predicted ones (147.2 nm for diameter, 0.099 for PDI and −36.17 mV for zeta potential), which proved that the mathematical model established by this method had a good predictability.
Nanoparticle Characterization
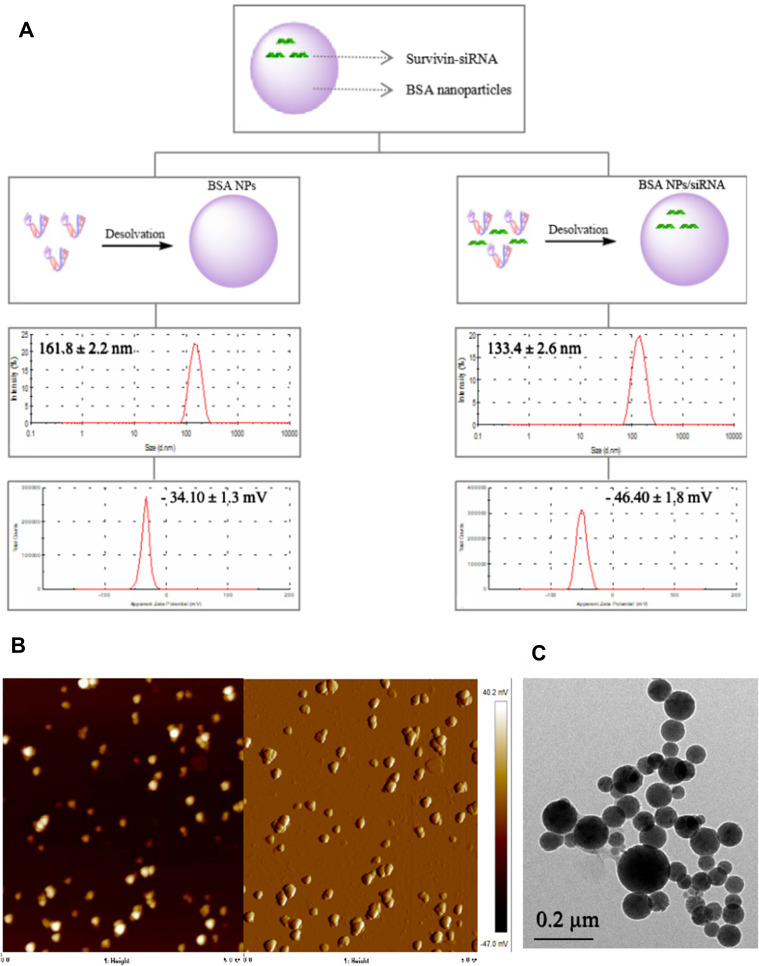
According to the optimal formula obtained above, BSA NPs/siRNA was preparation. The nanoparticles yield was calculated to be 93.1% ± 0.6%, and the survivin-siRNA loading efficiency of BSA NPs/siRNA was 0.85 ± 0.10 μg/mg. The hydrodynamic diameter and zeta potential of BSA NPs/siRNA were characterized by DLS. As shown in Figure 4A, the hydrodynamic diameter of BSA NPs/siRNA was 133.4 ± 2.6 nm, which was smaller than that of BSA NPs (161.8 ± 2.2 nm). And the zeta potential of BSA NPs/siRNA was −46.40 ± 1.8 mV, which was more negative than that of BSA NPs (−34.10 ± 1.3 mV).The potential mechanism for this phenomenon is as follows. siRNA may influence and compress the structure of BSA NPs, which makes the hydrodynamic diameter smaller,36 and the charge of siRNA is more negative when compared with BSA NPs, which makes the zeta potential of BSA NPs much more negative.24 The morphology of BSA NPs/siRNA was observed by AFM and TEM. As shown in Figure 4B and C, BSA NPs/siRNA had a spherical shape, and the size of BSA NPs/siRNA was uniformly distributed.
Figure 4.
Characteristics of BSA NPs and BSA NPs/siRNA. (A) The schematic diagram, hydrodynamic diameter and zeta potential for BSA NPs and BSA NPs/siRNA preparation, (B) AFM images of BSA NPs/siRNA, and (C) TEM image of BSA NPs/siRNA.
Nanoparticles Stability
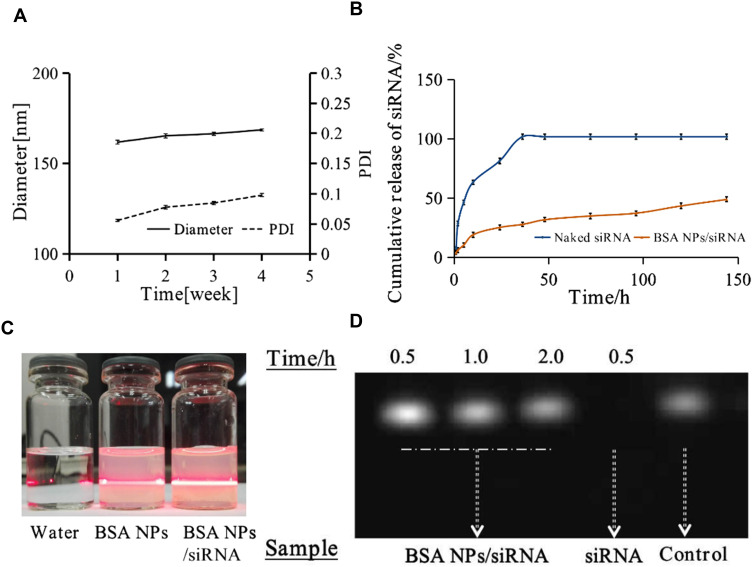
The storage stability of BSA NPs was studied at 4°C for 4 weeks. As shown in Figure 5A, the hydrodynamic diameter and polydispersity index of BSA NPs would not change greatly during the observation, which may attribute to the uniform particle size and negative zeta potential of BSA NPs/siRNA. As shown in Figure 5C, the Tyndall effect of water, BSA NPs and BSA NPs/siRNA was observed.
Figure 5.
(A) Storage stability of BSA NPs at 4°C for 4 weeks, which was characterized by diameter and PDI of BSA NPs. (B) The release of survivin-siRNA from naked siRNA and BSA NPs/siRNA. (C) The Tyndall effect of water, BSA NPs and BSA NPs/siRNA was observed. (D) The degradation of naked siRNA and BSA NPs protected siRNA by RNase A, which were characterized by agarose gel retardation assay. The data were presented as the mean ± SD, n = 3.
Degradation of siRNA by RNase in vivo is one of the biggest barriers for application of siRNA in clinical.37 So the gel retardation assay was applied to evaluate the protective effect of BSA NPs on siRNA degradation. As shown in Figure 5D, the gel strip of naked survivin-siRNA disappeared completely within 0.5 h. However, there was no attenuation for the strips of BSA NPs/siRNA even after 2 h. The results proved that BSA NPs could effectively protect siRNA from degradation by RNase A.
Release Profile of BSA NPs/siRNA
The release profile of BSA NPs/siRNA was measured using dialysis. As shown in Figure 5B, naked survivin-siRNA were totally released after 36 h when incubated at 37 °C, while only 28% ± 1.7% of survivin-siRNA was released from BSA NPs/siRNA at the same time. After incubation for 144 h, 49% ± 2.1% of survivin-siRNA was released from BSA NPs/siRNA. The results demonstrated that BSA NPs could maintain the sustained release of survivin-siRNA and was a potential controlled-release drug delivery system for siRNA.
Cellular Uptake
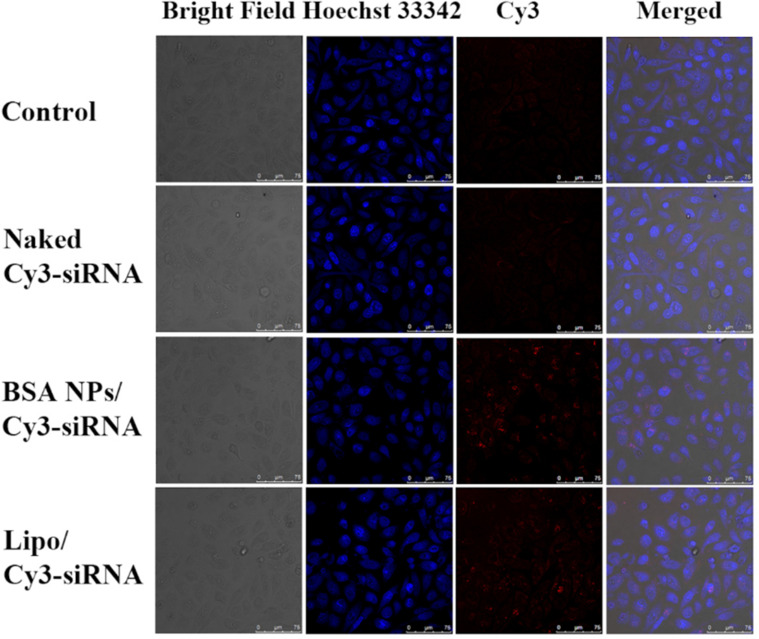
Cellular uptake of BSA NPs/siRNA was investigated on MCF-7 cells, and Cy3-labelled siRNA was applied to trace the position of siRNA in cells. As shown in Figure 6, the bright red fluorescence of Cy3-siRNA in BSA NPs/siRNA group was significantly stronger than that in naked siRNA group. The results suggested that naked siRNA could not be internalized by cells after incubation for 4 h, while BSA NPs/siRNA would deliver siRNA into cells efficiently.
Figure 6.
Cellular uptake of BSA NPs/siRNA in MCF-7 cells. The cells were treated with PBS (Control), Naked Cy3-siRNA, BSA NPs/Cy3-siRNA and Lipo/Cy3-siRNA for 4h, respectively, and the fluorescence signals were recorded by confocal laser scanning microscopy (CLSM). The blue fluorescence represented cell nucleus dyed by Hoechst 33342. The bright red fluorescence represented the location of Cy3-labelled siRNA.
In vitro Antiproliferation Assay
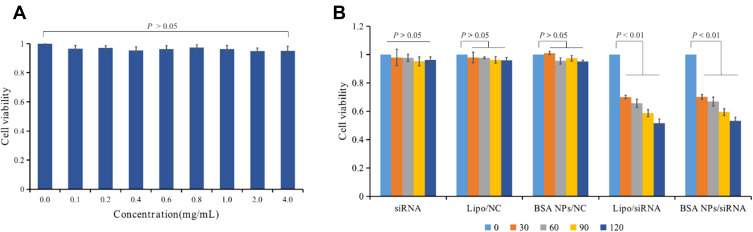
As reported, survivin-siRNA could inhibit cell proliferation by silencing of survivin protein.38–40 So the antiproliferation assay was performed to explore the inhibition effect of BSA NPs/siRNA on MCF-7 cells. Firstly, the cytotoxicity of BSA NPs was investigated by MTT assay. The results suggested that BSA NPs could not induce any toxicity for MCF-7 cells, and it was a safety vector for siRNA delivery (Figure 7A). And then, the antiproliferation effect of BSA NPs/siRNA was tested by MTT assay. As shown in Figure 7B, naked siRNA had no influence on cell viability at the tested concentrations (P > 0.05). Compared with naked siRNA, BSA NPs/siRNA decreased the viability of MCF-7 cells greatly, and the antiproliferation effect of BSA NPs/siRNA was enhanced significantly as the increase of siRNA concentrations (P < 0.01). Furthermore, BSA NPs/siRNA had almost the same inhibition effect as the positive control (Lipo/siRNA), which confirmed the excellent antitumor effect of BSA NPs/siRNA in vitro.
Figure 7.
(A) Cytotoxicity assay of BSA NPs tested by MTT method. (B) Anti-proliferation effect of siRNA, Lipo/NC, BSA NPs/NC, Lipo/siRNA and BSA NPs/siRNA at different concentrations on MCF-7 cells. The data were presented as the mean ± SD, n = 3.
Gene Silencing Effect of BSA NPs/siRNA
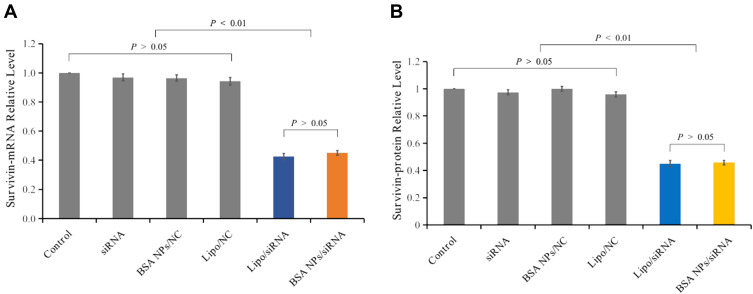
The in vitro gene silencing effect of BSA NPs/siRNA was investigated both on mRNA level and protein level. The downregulation of mRNA was measured by RT-PCR analysis. Compared with the control groups, BSA NPs/siRNA treated could decrease expression level of survivin-mRNA by 55% ± 1.6% (P < 0.01), which had no significant difference with the Lipo/siRNA-treated group (57% ± 2.2%, P > 0.05) (Figure 8A). These results confirmed that BSA NPs/siRNA could effectively silence the expression of survivin-mRNA by RNAi.
Figure 8.
Gene silencing effect of BSA NPs/siRNA in vitro. (A) Survivin-mRNA level of the MCF-7 cells treated with naked siRNA, Lipo/NC, BSA NPs/NC, Lipo/siRNA and BSA NPs/siRNA. (B) Survivin protein level of the MCF-7 cells treated with naked siRNA, Lipo/NC, BSA NPs/NC, Lipo/siRNA and BSA NPs/siRNA. The data were presented as the mean ± SD, n = 3.
The silencing effect in protein level was studied by ELISA analysis. Compared with the control groups, the protein downregulation rate for BSA NPs/siRNA was 54% ± 1.6% (P < 0.01), and the rate for Lipo/siRNA was 55% ± 2.3%. And there was no significant difference between the protein downregulation rates of BSA NPs/siRNA and Lipo/siRNA (P > 0.05) (Figure 8B). These results further proved that BSA NPs/siRNA could induce an efficient gene silencing effect by RNAi.
Distribution of BSA NPs/siRNA in vivo
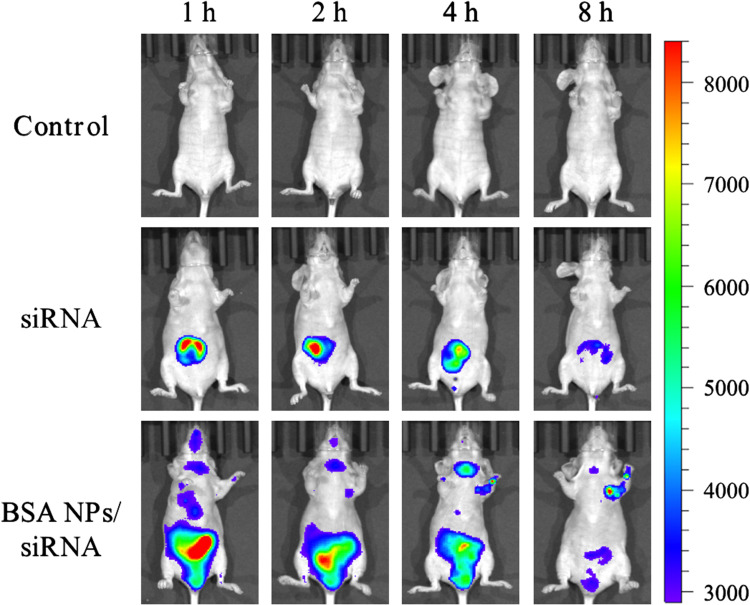
As it was reported, BSA NPs could accumulate in tumor tissues by passive and active targeting effect.41 Except for passive accumulation in tumor by EPR effect, BSA can bind with the albumin receptors (gp60 and SPARC) in tumor tissues to exert active tumor targeting effect.42 So the distribution of BSA NPs/siRNA was observed by fluorescent imaging on MCF-7 xenograft models. And Cy5 labelled survivin-siRNA was applied to trace the distribution of survivin-siRNA in vivo. As shown in Figure 9, there was a strong fluorescence in kidney of the survivin-siRNA-treated group at 1 h, and the fluorescent intensity was decreased with time. However, there was no fluorescence distributed in tumor tissues of the survivin-siRNA-treated group in the whole observation processes. As for BSA NPs/siRNA-treated group, the Cy5-labelled siRNA was mainly distributed in liver at 1h. As time increased, the fluorescence in liver decreased, and the fluorescence intensity of tumor enhanced gradually. These results suggested that BSA NPs would change the distribution of siRNA in vivo, and deliver siRNA into tumor tissues successfully by the targeting effect of BSA.
Figure 9.
Fluorescent images of tumor-bearing nude mice injected with normal saline, Cy5-siRNA and BSA NPs/Cy5-siRNA at 1 h, 2 h, 4 h and 8 h.
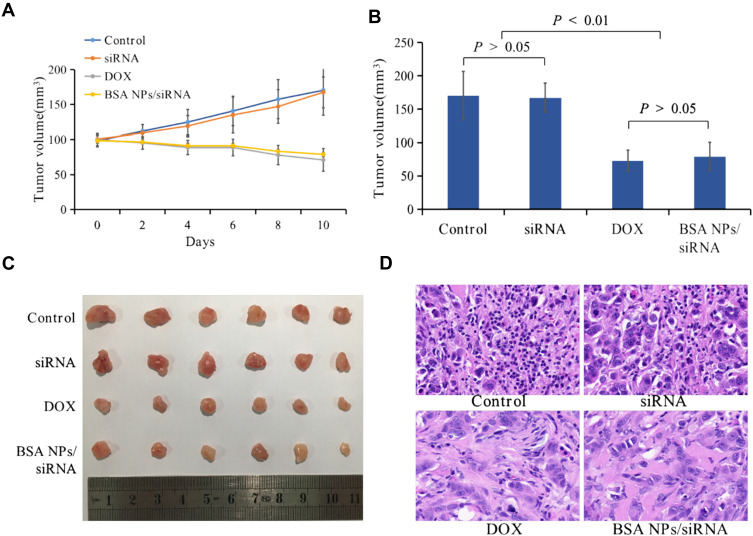
Tumor Inhibition of BSA NPs/siRNA in vivo
The tumor inhibition effect of BSA NPs/siRNA was also studied on MCF-7 xenograft models. Tumor volumes in naked siRNA group had a similar growth trend as that in control group, and there was no significant difference between the tumor volumes of naked siRNA and control group (Figure 10A, P > 0.05). However, in BSA NPs/siRNA group, the tumor volumes were decreased with time, and the tumor growth rate could be inhibited by 54% ± 12%. Moreover, the tumor inhibition effect of BSA NPs/siRNA was almost the same as that of DOX (the inhibition rate for DOX was 58 ± 9.6%, P > 0.05), confirming the excellent anti-tumor effect of BSA NPs/siRNA (Figure 10B and C). And the mechanism of BSA NPs/siRNA was investigated by H&E staining. As shown in Figure 10D, the tumor cells in control group and naked siRNA-treated group were closely arrange. However, in BSA NPs/siRNA-treated group, the tumor cells became sparse, and the intercellular space became large, which were the typical characters of necrosis. All of these results confirmed that BSA NPs/siRNA could effectively inhibit tumor growth by inducing cell apoptosis.
Figure 10.
In vivo antitumor effect of BSA NPs/siRNA. (A) The tumor volume of control, naked siRNA, DOX and BSA NPs/siRNA measured every other day. (B) The tumor volumes of control, naked siRNA, DOX and BSA NPs/siRNA at the end of the observation. (C) Tumor images of control, naked siRNA, DOX and BSA NPs/siRNA ex vivo. (D) The H&E staining of tumors in control, naked siRNA, DOX and BSA NPs/siRNA groups (40×). The data were presented as the mean ± SD, n=6.
Biosafety
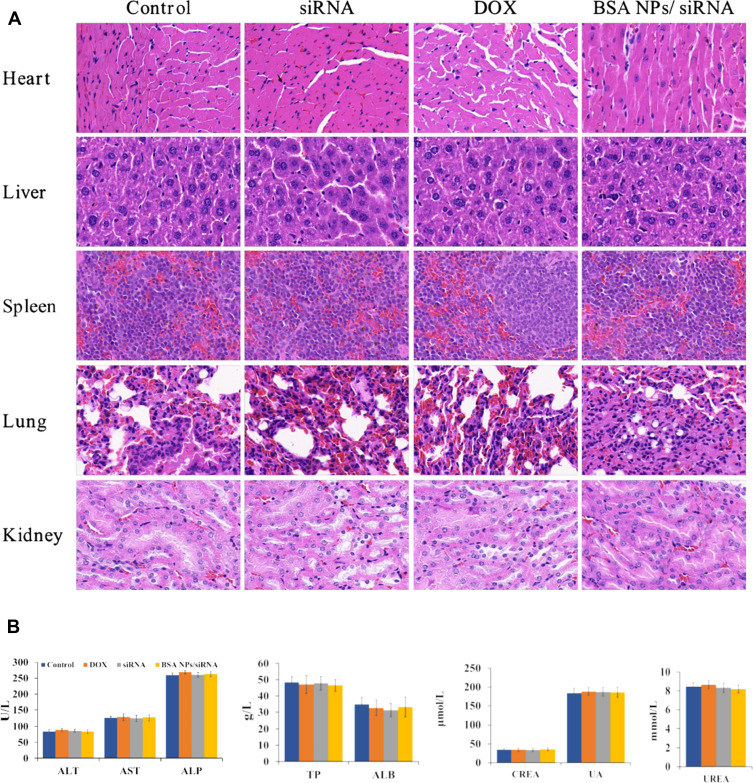
As an endogenous biomacromolecule, BSA has the advantages of biodegradability, non-toxicity, and non-immunogenicity.43 So it is hypothesized that BSA will have better biosafety as a gene delivery system, when compared with cationic materials. The biosafety of BSA NPs/siRNA in vivo was investigated by H&E stained histology test, blood panel analysis and blood biochemistry analysis. The H&E staining was applied to study the toxicity of BSA NPs/siRNA to the main organs. As shown in Figure 11A, the histological characters of the main organs (heart, liver, spleen, lung and kidney) in BSA NPs/siRNA-treated group had no significant differences with that in control group and naked siRNA-treated group. These results suggested that BSA NPs/siRNA had no damage to the main organs after administration in vivo. And the blood panel analysis was carried out to investigate the damages of BSA NPs/siRNA to the blood parameters (Table 2). The results suggested that there was no significant difference between BSA NPs/siRNA-treated group and the control groups as for hematology analysis. Furthermore, the blood biochemistry analysis was carried out to study the damages of BSA NPs/siRNA to the function of livers and kidneys. The liver function indexes (ALT, AST, ALP, TP and ALB) and the kidney function indexes (CREA, UA and UREA) were tested. As shown in Figure 11B, no significant differences were noted between BSA NPs/siRNA-treated group and the control groups, which proved that BSA NPS/siRNA was safety to the function of livers and kidneys. All of the results above suggested that the BSA NPs/siRNA was a biosafety material for the delivery of siRNA, and would not induce any organic or blood damages after administration in vivo.
Figure 11.
Biosafety of BSA NPs/siRNA. (A) H&E staining of the main organs (hearts, livers, spleens, lungs and kidneys) treated by NS (control), naked siRNA, DOX and BSA NPs/siRNA (40×). (B) Blood biochemistry indexes of NS (control), naked siRNA, DOX and BSA NPs/siRNA-treated groups. ALT, AST, ALP, TP and ALB indicated the function of liver; CREA, UA and UREA indicated the function of kidney. The data were presented as the mean ± SD, n=5.
Table 2.
Blood Panel Analysis
| Unit | Control | DOX | siRNA | BSA NPs/siRNA | |
|---|---|---|---|---|---|
| WBC | 109/L | 3.82 ± 0.42 | 4.10 ± 0.49 | 3.86 ± 0.55 | 3.62 ± 0.32 |
| RBC | 1012/L | 9.07 ± 0.57 | 8.41 ± 1.06 | 9.17 ± 0.83 | 9.21 ± 0.74 |
| HGB | g/L | 139.40 ± 8.96 | 131.06 ± 13.32 | 138.60 ± 7.50 | 133.80 ± 7.73 |
| HCT | % | 42.62 ± 2.58 | 41.40 ± 6.32 | 42.92 ± 2.88 | 42.54 ± 3.89 |
| MCV | fL | 53.98 ± 2.18 | 53.58 ± 2.52 | 54.60 ± 2.46 | 53.14 ± 2.68 |
| MCH | pg | 17.86 ± 0.32 | 17.52 ± 0.70 | 17.46 ± 0.29 | 17.90 ± 0.56 |
| MCHC | g/L | 331.32 ± 6.21 | 325.60 ± 8.68 | 329.00 ± 14.68 | 331.04 ± 10.95 |
| PLT | 109/L | 585.34 ± 15.29 | 597.80 ± 37.76 | 591.40 ± 30.02 | 586.18 ± 27.55 |
| RDW | % | 21.94 ± 1.72 | 22.92 ± 2.98 | 22.30 ± 2.41 | 23.06 ± 2.85 |
Note: The data were presented as the mean ± SD, n=5.
Abbreviations: WBC, white blood cell; RBC, red blood cell; HGB, hemoglobin; HCT, hematocrit; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; RDW, red blood cell distribution width; PLT, platelet.
Discussion
Because of the excellent biocompatibility, non-toxicity and non-immunogenicity of bovine serum albumin, BSA NPs have been widely designed as a drug carrier system for delivery of chemical drugs, gene drugs and so on.44 It is critical to optimize the preparation to obtain BSA NPs with the best characters as a drug delivery system.
In this research, a prescription optimization method CCD-RSM was applied to get the optimal formula for BSA NPs preparation.45 And the optimal formula was settled to be 20 mg/mL for BSA concentration, 9 for pH value, 136% for crosslinking degree and 1.6mL/min for speed of ethanol addition. Moreover, the diameter, PDI and zeta potential of BSA NPs prepared by the optimal formula were the same as these predicted ones, which proved that the mathematical model established by this method had a good predictability. And the optimal formula was carried out to prepare survivin-siRNA loaded BSA NPs. The hydrodynamic diameter of BSA NPs/siRNA was smaller than that of BSA NPs, and the zeta potential of BSA NPs/siRNA was more negative than that of BSA NPs. It is speculated that siRNA could condense BSA NPs, and also proved that survivin-siRNA was loaded into BSA NPs successfully.
The in vitro research suggested that BSA NPs could not only enhance the stability of survivin-siRNA but also promote the uptake of survivin-siRNA to increase its anti-tumor effect in vitro. The stability study suggested that BSA NPs/siRNA could remain stable at 4°C for 4 weeks, and could protect survivin-siRNA from degradation by RNase A. The release profile of BSA NPs/siRNA demonstrated that BSA NPs/siRNA could maintain a sustained release of siRNA. The cellular uptake results confirmed that naked siRNA could not be internalized by cells in the course of observation, while BSA NPs/siRNA could deliver siRNA into cells efficiently. The brilliant siRNA delivery of BSA NPs/siRNA may be attributed to the dual targeting ability of BSA by gp60 and SPARC.46 Considering the results above, BSA NPs/siRNA would have an excellent anti-tumor effect in vitro. The in vitro antiproliferation assay proved that BSA NPs/siRNA had an equal antitumor effect as Lipo/siRNA by downregulating the expression of survivin-mRNA and survivin protein.
The in vivo research suggested that BSA NPs could change the distribution of siRNA by targeting effect, so as to inhibit tumor growth in vivo. The in vivo imaging experiments suggested that the fluorescence intensity in tumor tissues was stronger in BSA NPs/siRNA-treated groups. However, more fluorescence was observed in kidney in naked siRNA-treated group, which confirmed that BSA NPs could protect siRNA from clearance by kidney. It was reported that BSA could bind with the albumin receptors (gp60 and SPARC) in tumor tissues to exert active tumor targeting effect, so the distribution of BSA NPs/siRNA in tumors may contribute to the passive and active targeting effect of BSA NPs.47,48 And the in vivo antitumor study suggested that the tumor inhibition rate of BSA NPs/siRNA was much higher than naked siRNA, which was almost the same as that of DOX. Besides, the H&E staining assay, the blood panel and the blood biochemistry analysis proved that BSA NPs was a biosafety material for siRNA delivery, due to the brilliant biocompatibility of BSA.49
In summary, CCD-RSM is an effective tool for preparation analysis, and the optimal formula obtained by this method had a good predictability for BSA NPs preparation. What’s more, BSA NPs/siRNA prepared by the optimal formula was an attractive gene delivery system, and it could enhance the gene silencing effect of survivin-siRNA by promoting cellular uptake and tumor targeting effect.
Conclusions
In summary, a BSA-based nanoparticle was prepared for the delivery of survivin-siRNA in this research. In order to obtain the optimal formula for the preparation of BSA NPs, the multi-factor experiments were designed by CCD-RSM. And the optimal formula was applied for the preparation of BSA NPs/siRNA, which had a brilliant antitumor and biocompatibility both in vitro and in vivo. The BSA NPs/siRNA could remain the stability of siRNA, and promote the uptake of siRNA by MCF-7 cells, which ensured the excellent anti-tumor effect of survivin-siRNA in vitro. Moreover, after loaded in BSA NPs, more survivin-siRNA was distributed in tumor tissues after administration in vivo, due to the passive and active targeting effect. So BSA NPs/siRNA had an impressive tumor inhibition effect in vivo. Besides, the toxicity and biosafety research suggested that BSA NPs was a safety material for siRNA delivery. In summary, BSA NPs prepared by the optimal formula was a promising vector for tumor-targeting gene delivery. This research provides an efficient method for design of BSA NPs delivery system, and also depicts an excellent vector with remarkable gene delivery ability.
Acknowledgments
The authors gratefully acknowledge the support from Beijing Area Major Laboratory of Peptide and Small Molecular Drugs, Engineering Research Center of Endogenous Prophylactic of Ministry of Education of China, and Beijing Laboratory of Biomedical Materials.
Funding Statement
This work was supported by the National Natural Science Foundation (81502688), Beijing Natural Science Foundation Program and Scientific Research Key Program of Beijing Municipal Commission of Education (KM201810025019), and a basic-clinical key research grant (16JL72, 17JL67) from Capital Medical University, the Importation and Development of High-Caliber Talents Project of Beijing Municipal Institutions (2013–2015).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest.
References
- 1.Kim B, Park JH, Sailor MJ. Rekindling RNAi therapy: materials design requirements for in vivo siRNA delivery. Adv Mater. 2019;31(49):e1903637. doi: 10.1002/adma.201903637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yonezawa S, Koide H, Asai T. Recent advances in siRNA delivery mediated by lipid-based nanoparticles. Adv Drug Deliv Rev. 2020;154–155:64–78. doi: 10.1016/j.addr.2020.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- 4.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15(2):188–200. doi: 10.1101/gad.862301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulkarni JA, Witzigmann D, Chen S, Cullis PR, van der Meel R. Lipid nanoparticle technology for clinical translation of siRNA therapeutics. Acc Chem Res. 2019;52(9):2435–2444. doi: 10.1021/acs.accounts.9b00368 [DOI] [PubMed] [Google Scholar]
- 6.Gavrilov K, Saltzman WM. Therapeutic siRNA: principles, challenges, and strategies. Yale J Biol Med. 2012;85(2):187–200. [PMC free article] [PubMed] [Google Scholar]
- 7.Akinc A, Maier MA, Manoharan M, et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat Nanotechnol. 2019;14(12):1084–1087. doi: 10.1038/s41565-019-0591-y [DOI] [PubMed] [Google Scholar]
- 8.Akbaba H, Erel-Akbaba G, Kotmak M, Baspinar Y. Enhanced cellular uptake and gene silencing activity of survivin-siRNA via ultrasound-mediated nanobubbles in lung cancer cells. Pharm Res. 2020;37(8):165. doi: 10.1007/s11095-020-02885-x [DOI] [PubMed] [Google Scholar]
- 9.Baspinar Y, Erel-Akbaba G, Kotmak M, Akbaba H. Development and characterization of nanobubbles containing paclitaxel and survivin inhibitor YM155 against lung cancer. Int J Pharm. 2019;566:149–156. doi: 10.1016/j.ijpharm.2019.05.039 [DOI] [PubMed] [Google Scholar]
- 10.Dong H, Yao L, Bi W, Wang F, Song W, Lv Y. Combination of survivin siRNA with neoadjuvant chemotherapy enhances apoptosis and reverses drug resistance in breast cancer MCF-7 cells. J Cancer Res Ther. 2015;11(4):717–722. doi: 10.4103/0973-1482.147764 [DOI] [PubMed] [Google Scholar]
- 11.Elzoghby AO, Samy WM, Elgindy NA. Albumin-based nanoparticles as potential controlled release drug delivery systems. J Control Release. 2012;157(2):168–182. doi: 10.1016/j.jconrel.2011.07.031 [DOI] [PubMed] [Google Scholar]
- 12.Bhushan B, Khanadeev V, Khlebtsov B, Khlebtsov N, Gopinath P. Impact of albumin based approaches in nanomedicine: imaging, targeting and drug delivery. Adv Colloid Interface Sci. 2017;246:13–39. doi: 10.1016/j.cis.2017.06.012 [DOI] [PubMed] [Google Scholar]
- 13.Wilson B, Ambika TV, Patel RD, Jenita JL, Priyadarshini SR. Nanoparticles based on albumin: preparation, characterization and the use for 5-flurouracil delivery. Int J Biol Macromol. 2012;51(5):874–878. doi: 10.1016/j.ijbiomac.2012.07.014 [DOI] [PubMed] [Google Scholar]
- 14.Ruan C, Liu L, Lu Y, et al. Substance P-modified human serum albumin nanoparticles loaded with paclitaxel for targeted therapy of glioma. Acta Pharm Sin B. 2018;8(1):85–96. doi: 10.1016/j.apsb.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sleep D. Albumin and its application in drug delivery. Expert Opin Drug Deliv. 2015;12(5):793–812. doi: 10.1517/17425247.2015.993313 [DOI] [PubMed] [Google Scholar]
- 16.Kratz F. Albumin as a drug carrier: design of prodrugs, drug conjugates and nanoparticles. J Control Release. 2008;132(3):171–183. doi: 10.1016/j.jconrel.2008.05.010 [DOI] [PubMed] [Google Scholar]
- 17.Arnedo A, Espuelas S, Irache JM. Albumin nanoparticles as carriers for a phosphodiester oligonucleotide. Int J Pharm. 2002;244(1–2):59–72. doi: 10.1016/S0378-5173(02)00300-9 [DOI] [PubMed] [Google Scholar]
- 18.Li L, Zhao X, Yang C, Hu H, Qiao M, Chen D. Preparation and optimization of doxorubicin-loaded albumin nanoparticles using response surface methodology. Drug Dev Ind Pharm. 2011;37(10):1170–1180. doi: 10.3109/03639045.2011.563781 [DOI] [PubMed] [Google Scholar]
- 19.Tsai S, Ting Y. Synthesize of alginate/chitosan bilayer nanocarrier by CCD-RSM guided co-axial electrospray: a novel and versatile approach. Food Res Int. 2019;116:1163–1172. doi: 10.1016/j.foodres.2018.11.047 [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Feng JF, Zhang H, Luo JY. Optimization preparation of chansu-loaded solid lipid nanoparticles by central composite design and response surface method. Zhongguo Zhong Yao Za Zhi. 2006;31(8):650–653. [PubMed] [Google Scholar]
- 21.Liu M, Chen JH, Dong FR, Liu Y. Optimized preparation of ginkgolides A and B long-circulating solid lipid nanoparticles by central composite design and response surface method. Nan Fang Yi Ke Da Xue Xue Bao. 2008;28(5):700–703. [PubMed] [Google Scholar]
- 22.Ding L, Liu L, Xu D. Optimized preparation of dry powder inhalation of Rehmannia glutinosa oligosaccharides by central composite design and response surface method. Zhongguo Zhong Yao Za Zhi. 2009;34(24):3203–3206. [PubMed] [Google Scholar]
- 23.Irache JM, Merodio M, Arnedo A, Camapanero MA, Mirshahi M, Espuelas S. Albumin nanoparticles for the intravitreal delivery of anticytomegaloviral drugs. Mini Rev Med Chem. 2005;5(3):293–305. doi: 10.2174/1389557053175335 [DOI] [PubMed] [Google Scholar]
- 24.Choi JH, Hwang HJ, Shin SW, Choi JW, Um SH, Oh BK. A novel albumin nanocomplex containing both small interfering RNA and gold nanorods for synergetic anticancer therapy. Nanoscale. 2015;7(20):9229–9237. doi: 10.1039/C5NR00211G [DOI] [PubMed] [Google Scholar]
- 25.Song CX, Chen HM, Dai Y, Kang M, Hu J, Deng Y. Optimization of process of icraiin be hydrolyzed to Baohuoside I by cellulase based on Plackett-Burman design combined with CCD response surface methodology. Zhong Yao Cai. 2014;37(11):2082–2086. [PubMed] [Google Scholar]
- 26.Nicolás P, Lassalle VL, Ferreira ML. Quantification of immobilized Candida antarctica lipase B (CALB) using ICP-AES combined with Bradford method. Enzyme Microb Technol. 2017;97:97–103. doi: 10.1016/j.enzmictec.2016.11.009 [DOI] [PubMed] [Google Scholar]
- 27.Arnedo A, Campanero MA, Espuelas S, Renedo MJ, Irache JM. Determination of oligonucleotide ISIS 2922 in nanoparticulate delivery systems by capillary zone electrophoresis. J Chromatogr A. 2000;871(1–2):311–320. doi: 10.1016/S0021-9673(99)01193-0 [DOI] [PubMed] [Google Scholar]
- 28.Mehta A, Dalle Vedove E, Isert L, Merkel OM. Targeting KRAS mutant lung cancer cells with siRNA-loaded bovine serum albumin nanoparticles. Pharm Res. 2019;36(9):133. doi: 10.1007/s11095-019-2665-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Sun Q, Cui C, Li J, Wang Y. Anti-HER2 functionalized graphene oxide as survivin-siRNA delivery carrier inhibits breast carcinoma growth in vitro and in vivo. Drug Des Devel Ther. 2018;12:2841–2855. doi: 10.2147/DDDT.S169430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Q, Wang X, Cui C, Li J, Wang Y. Doxorubicin and anti-VEGF siRNA co-delivery via nano-graphene oxide for enhanced cancer therapy in vitro and in vivo. Int J Nanomedicine. 2018;13:3713–3728. doi: 10.2147/IJN.S162939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bi Y, Zhang Y, Cui C, Ren L, Jiang X. Gene-silencing effects of anti-survivin siRNA delivered by RGDV-functionalized nanodiamond carrier in the breast carcinoma cell line MCF-7. Int J Nanomedicine. 2016;11:5771–5787. doi: 10.2147/IJN.S117611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Ge X, Cui C, et al. Preparation and characterization of functionalized graphene oxide carrier for siRNA delivery. Int J Mol Sci. 2018;19(10):3202. doi: 10.3390/ijms19103202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui C, Wang Y, Zhao W, et al. RGDS covalently surfaced nanodiamond as a tumor targeting carrier of VEGF-siRNA: synthesis, characterization and bioassay. J Mater Chem B. 2015;3(48):9260–9268. doi: 10.1039/C5TB01602A [DOI] [PubMed] [Google Scholar]
- 34.Son S, Song S, Lee SJ, et al. Self-crosslinked human serum albumin nanocarriers for systemic delivery of polymerized siRNA to tumors. Biomaterials. 2013;34(37):9475–9485. doi: 10.1016/j.biomaterials.2013.08.085 [DOI] [PubMed] [Google Scholar]
- 35.Yang T, Li B, Qi S, et al. Co-delivery of doxorubicin and Bmi1 siRNA by folate receptor targeted liposomes exhibits enhanced anti-tumor effects in vitro and in vivo. Theranostics. 2014;4(11):1096–1111. doi: 10.7150/thno.9423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnedo A, Irache JM, Merodio M, Espuelas Millán MS. Albumin nanoparticles improved the stability, nuclear accumulation and anticytomegaloviral activity of a phosphodiester oligonucleotide. J Control Release. 2004;94(1):217–227. doi: 10.1016/j.jconrel.2003.10.009 [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Ye YF. Effect of survivin siRNA on biological behaviour of breast cancer MCF7 cells. Asian Pac J Trop Med. 2015;8(3):225–228. doi: 10.1016/S1995-7645(14)60320-5 [DOI] [PubMed] [Google Scholar]
- 38.Larsen MT, Kuhlmann M, Hvam ML, Howard KA. Albumin-based drug delivery: harnessing nature to cure disease. Mol Cell Ther. 2016;4:3. doi: 10.1186/s40591-016-0048-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parodi A, Miao J, Soond SM, Rudzińska M, Zamyatnin AA. Albumin nanovectors in cancer therapy and imaging. Biomolecules. 2019;9(6):218. doi: 10.3390/biom9060218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kandav G, Bhatt DC, Jindal DK, Singh SK. Formulation, optimization, and evaluation of allopurinol-loaded bovine serum albumin nanoparticles for targeting kidney in management of hyperuricemic nephrolithiasis: formulation, optimization, and evaluation of ABNPs for kidney targeting. AAPS PharmSciTech. 2020;21(5):164. doi: 10.1208/s12249-020-01695-z [DOI] [PubMed] [Google Scholar]
- 41.Langer K, Balthasar S, Vogel V, Dinauer N, von Briesen H, Schubert D. Optimization of the preparation process for human serum albumin (HSA) nanoparticles. Int J Pharm. 2003;257(1–2):169–180. doi: 10.1016/S0378-5173(03)00134-0 [DOI] [PubMed] [Google Scholar]
- 42.Langer K, Anhorn MG, Steinhauser I, et al. Human serum albumin (HSA) nanoparticles: reproducibility of preparation process and kinetics of enzymatic degradation. Int J Pharm. 2008;347(1–2):109–117. doi: 10.1016/j.ijpharm.2007.06.028 [DOI] [PubMed] [Google Scholar]
- 43.Liu X, Mohanty RP, Maier EY, et al. Controlled loading of albumin-drug conjugates ex vivo for enhanced drug delivery and antitumor efficacy. J Control Release. 2020;328:1–12. doi: 10.1016/j.jconrel.2020.08.015 [DOI] [PubMed] [Google Scholar]
- 44.Loureiro A, Azoia NG, Gomes AC, Cavaco-Paulo A. Albumin-based nanodevices as drug carriers. Curr Pharm Des. 2016;22(10):1371–1390. doi: 10.2174/1381612822666160125114900 [DOI] [PubMed] [Google Scholar]
- 45.Lomis N, Westfall S, Farahdel L, Malhotra M, Shum-Tim D, Prakash S. Human serum albumin nanoparticles for use in cancer drug delivery: process optimization and in vitro characterization. Nanomaterials. 2016;6(6):116. doi: 10.3390/nano6060116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarhini M, Pizzoccaro A, Benlyamani I, et al. Human serum albumin nanoparticles as nanovector carriers for proteins: application to the antibacterial proteins “neutrophil elastase” and “secretory leukocyte protease inhibitor”. Int J Pharm. 2020;579:119–150. doi: 10.1016/j.ijpharm.2020.119150 [DOI] [PubMed] [Google Scholar]
- 47.Yogasundaram H, Bahniuk MS, Singh HD, Aliabadi HM, Uludağ H, Unsworth LD. BSA nanoparticles for siRNA delivery: coating effects on nanoparticle properties, plasma protein adsorption, and in vitro siRNA delivery. Int J Biomater. 2012;2012:1–10. doi: 10.1155/2012/584060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elsadek B, Kratz F. Impact of albumin on drug delivery–new applications on the horizon. J Control Release. 2012;157(1):4–28. doi: 10.1016/j.jconrel.2011.09.069 [DOI] [PubMed] [Google Scholar]
- 49.Singh HD, Wang G, Uluda H, Unsworth LD. Poly-L-lysine-coated albumin nanoparticles: stability, mechanism for increasing in vitro enzymatic resilience, and siRNA release characteristics. Acta Biomater. 2010;6(11):4277–4284. doi: 10.1016/j.actbio.2010.06.017 [DOI] [PubMed] [Google Scholar]