Fig. 2.
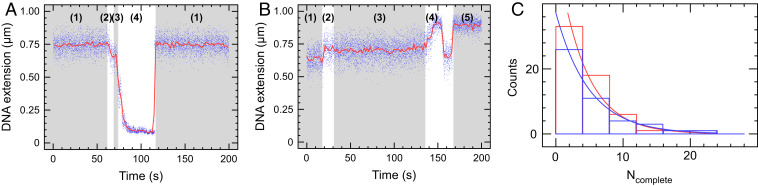
Translocase-deficient MfdRA953 maintains the ability to form tripartite supercoiled domain. (A) Time-trace showing phase (1) positively supercoiled DNA, phase (2) RNAP initiation and phase (3) elongation. In phase (4), MfdRA953 binds to elongating RNAP and DNA to form the tripartite supercoiled domain. As RNAP terminates, the domains dissolve and annihilate, and the DNA returns to the phase (1) baseline extension. (B) Time-trace showing phase (1) negatively supercoiled DNA, phase (2) RNAP initiation, and phase (3) elongation. These signals are in the opposing direction to A because introduction of positive supercoiling into negatively supercoiled DNA leads to an increase in DNA extension. In phase (4), MfdRA953 binds to elongating RNAP and DNA to form the tripartite supercoiled domain. The gain of positive supercoils first annihilates the negative supercoils (DNA extension increases) before reaching the maximal extension state, after which point there is a net gain of positive supercoils which causes the bead to descend toward the surface. When RNAP terminates or Mfd dissociates from either RNAP or DNA, in phase (5) a new high-extension intermediate state is observed for negatively supercoiled DNA substrate only. (C) The frequency distribution of tripartite supercoiled domain events is fitted to an exponential distribution. Ncomplete is defined for each DNA molecule as the number of uninterrupted RNAP transcription cycles completed on that given DNA molecule before a tripartite supercoiled domain event interrupts a transcription cycle; a low Ncomplete indicates an elevated frequency of transcription events displaying tripartite supercoiled domain formation. With 50 nM GreB (blue), the average of Ncomplete is 4.2 ± 0.6 (SEM, n = 59) and without GreB (red) the average of Ncomplete is 5.0 ± 0.9 (SEM, n = 46).