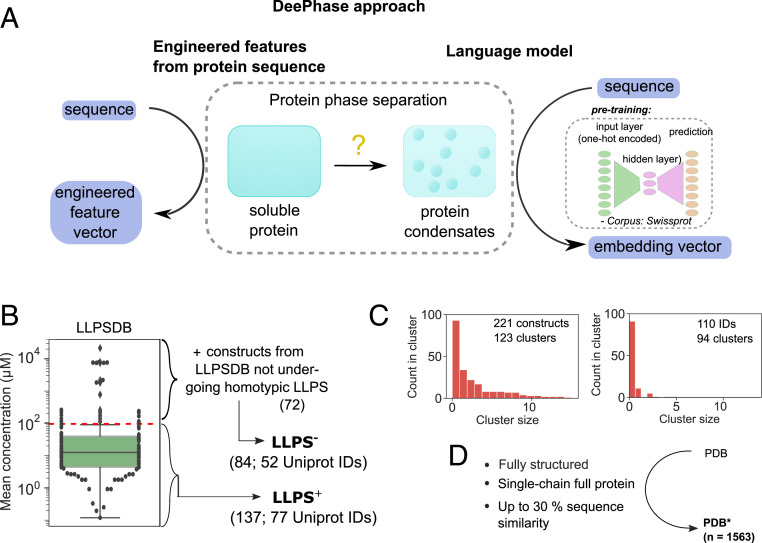
Fig. 1.
(A) DeePhase predicts the propensity of proteins to undergo phase separation by combining engineered features computed directly from protein sequences with protein sequence embedding vectors generated using a pretrained language model. The DeePhase model was trained using three datasets, namely two classes of intrinsically disordered proteins with a different LLPS propensity ( and ) and a set of structured sequences (). (B) To generate the and datasets, the entries in the LLPSDB database (27) were filtered for single-protein systems. The constructs that phase separated at an average concentration below were classified as having a high LLPS propensity (; 137 constructs from 77 UniProt IDs) with the remaining 25 constructs together with constructs that had not been observed to phase separate homotypically classified as low-propensity dataset (; 84 constructs from 52 UniProt IDs). (C) The 221 sequences clustered into 123 different clusters [Left, CD-hit clustering algorithm (28) with the lowest threshold of 0.4]. (Right) The 110 parent sequences showed high diversity by forming 94 distinct clusters. (D) The dataset (1,563 constructs) was constructed by filtering the entries in the PDB (29) to fully structured full-protein single chains and clustering for sequence similarity with a single entry selected from each cluster.