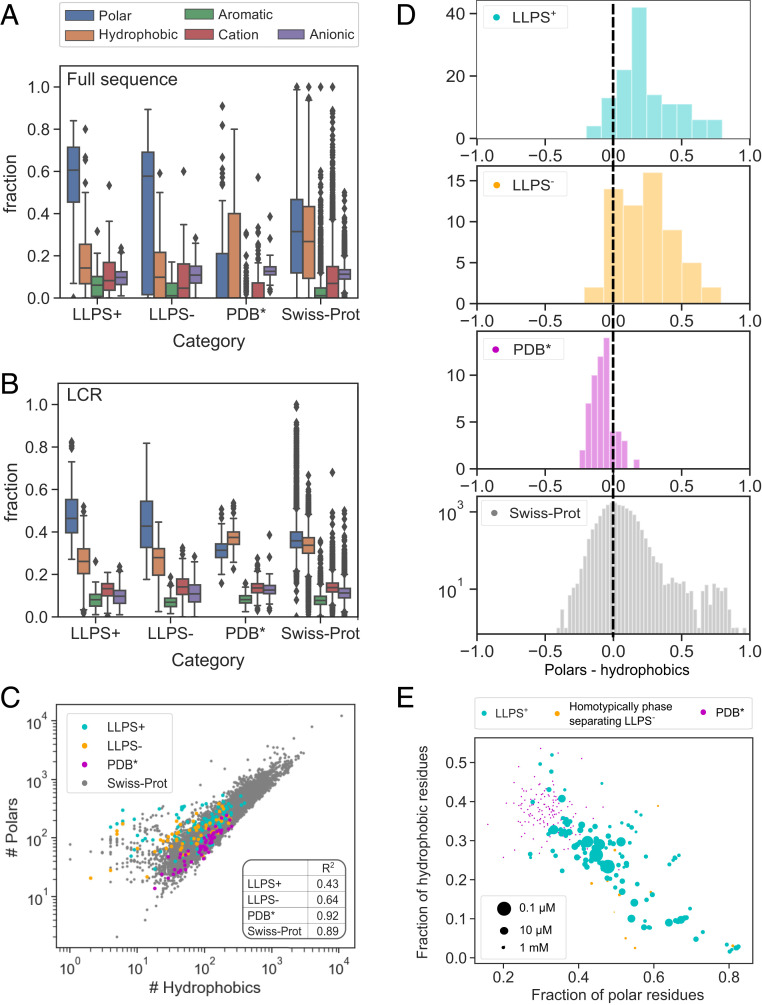
Fig. 3.
Comparison of the amino acid composition of the sequences within the , , , and Swiss-Prot datasets. (A) LLPS-prone sequences were enriched in polar amino acid residues (blue) and depleted in hydrophobic (orange) and anionic (purple) residues. (B) The elevated abundance of polar residues was particularly pronounced within the LCR with the sequences in the also having their LCR regions enriched in aromatic (green) and cationic (red) residues compared to in the Swiss-Prot database. (C) The relationship between polar and hydrophobic residues was less tightly conserved among LLPS-prone sequences as indicated by smaller values (Inset). (D) Compared to the Swiss-Prot and the datasets, LLPS-prone proteins showed an overabundance of polar residues relative to hydrophobic ones. (E) When further examining the relationship between the saturation concentration of a construct (indicated by the marker size; saturation concentration of 10 mM was assumed for ) and its amino acid composition, a strong overabundance of polar residues was seen to lead to an increase in saturation concentration with most LLPS-prone sequences showing a tight balance between the abundances of polar and hydrophobic residues.