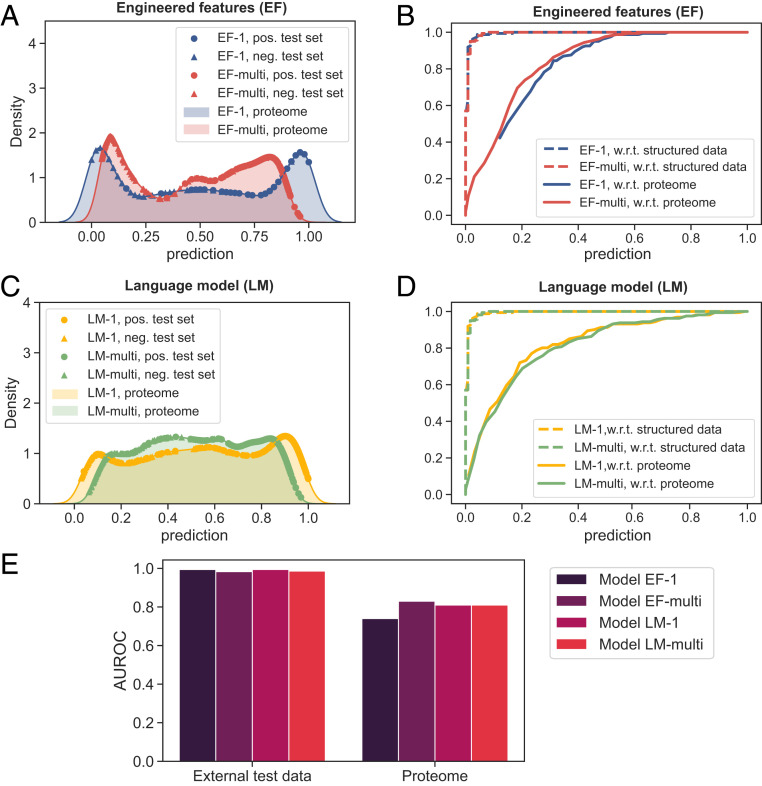
Fig. 5.
Performance of the models on external data when 1) discriminating between LLPS-prone sequences and structured proteins and 2) identifying LLPS-prone proteins from the human proteome. (A) The prediction profiles of model EF-1 (trained on and ) and model EF-multi (trained on all three protein classes) on external test data comprising 161 LLPS-prone sequences (pos.; colored circles) and 161 sequences highly unlikely to undergo phase separation (neg.; colored triangles) and the human proteome (colored region; 20,291 proteins). The positive part of the external dataset was constructed based on the PhaSepDB database with sequences that had their Uniprot IDs overlapping with the training data excluded. The negative half was based on the . (B) ROC curves of the models 1) on the external test data (dashed line) and 2) when identifying LLPS-prone sequences from the human proteome by regarding all proteins that had not been reported to phase separate as nonphase separating (lower bound for the false positive rate; solid line). (C and D) Same data for models LM-1 and LM-multi where 200-dimensional representations learned from a pretrained word2vec model were used for featurization. (E) Comparison of the AUROC values for ROC curves shown in B and D for the two tasks.