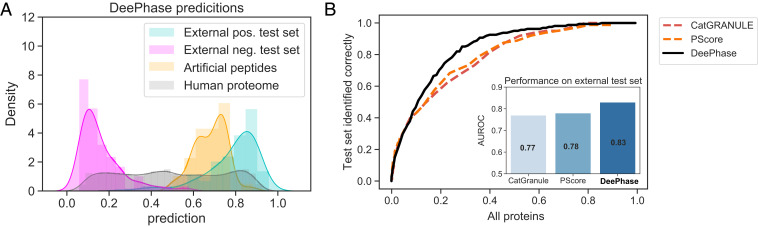
Fig. 7.
Generalizability of DeePhase to evolutionarily nonrelated sequences and comparison to previously developed LLPS predictors. (A) DeePhase prediction profile on the human proteome (gray), on the external test data (161 LLPS-prone [cyan] and 161 structured proteins [pink]), and on a set of 73 artificial peptides that have been experimentally validated to be phase separate (38) (yellow). DeePhase allocated a high LLPS-propensity score for the latter dataset, indicating that its capability to evaluate phase behavior extends to evolutionary nonrelated sequences. (B) Comparison of DeePhase to CatGRANULE and PScore, the algorithms that were recently found to be the best performing for LLPS prediction (23), when identifying LLPS-prone sequences from the human proteome. For a reliable comparison, sequences were filtered for a length of 140 residues or above as this is the lowest threshold at which the PScore can be evaluated.