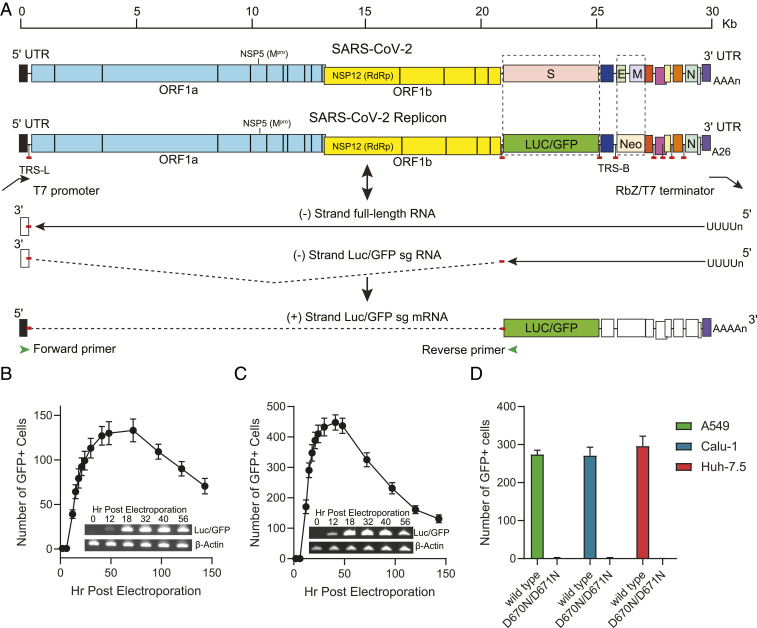
Fig. 1.
Construction and transcriptional analysis of SARS-CoV-2 replicon. (A) Schematic diagram of reporter replicon genome structure, replication, and sg mRNA production. The S and E/M ORFs in the wt viral genome were deleted and replaced with Luc/GFP and Neo genes, respectively. The full-length replicon cDNA was flanked by a T7 promoter and a polyA/HDV ribozyme and T7 terminator cassette. The in vitro-transcribed replicon RNA is copied by replicase complex to produce genomic or subgenomic-sized negative stranded RNAs (solid lines). The negative stranded sg RNAs are used as templates to produce the sg mRNAs for expression of reporter and structural proteins. The sg mRNAs consist of the leader at 5′-UTR of the genome and mRNA body sequences joined by a short and conserved sequence motif, the transcription-regulating sequence (TRS). The location of the sequences encoding major antiviral targets NSP5 (Mpro) and NSP12 (RdRp), the untranscribed regions (dashed lines), TRS leader (TRS-L) and TRS body (TRS-B) are also indicated. (B and C) Kinetics of GFP expression post-electroporation and detection of the sg mRNA expressing Luc/GFP reporter products. 293T (B) and A549 (C) cells were electroporated with in vitro-transcribed replicon RNA, and the number of GFP-positive cells in each well containing 1 × 104 cells were counted at indicated time points. Total RNA was also collected and purified at indicated times from electroporated cells. RT-PCR products from mRNAs expressing Luc/GFP or β-actin were confirmed by gel electrophoresis. (D) GFP reporter signals produced by the wt and RdRp mutant replicons. The A549, Calu-1, and Huh-7.5 cells were electroporated with T7 polymerase transcribed wt replicon RNA or the RdRp D760N/D761N double-mutant replicon RNA and transferred to 384-well plates. The number of GFP-positive cells in each well were counted at 30 h post-electroporation.