Figure 1.
hsa_circ_0001018 was highly expressed in PTC tissues and PTC cell lines
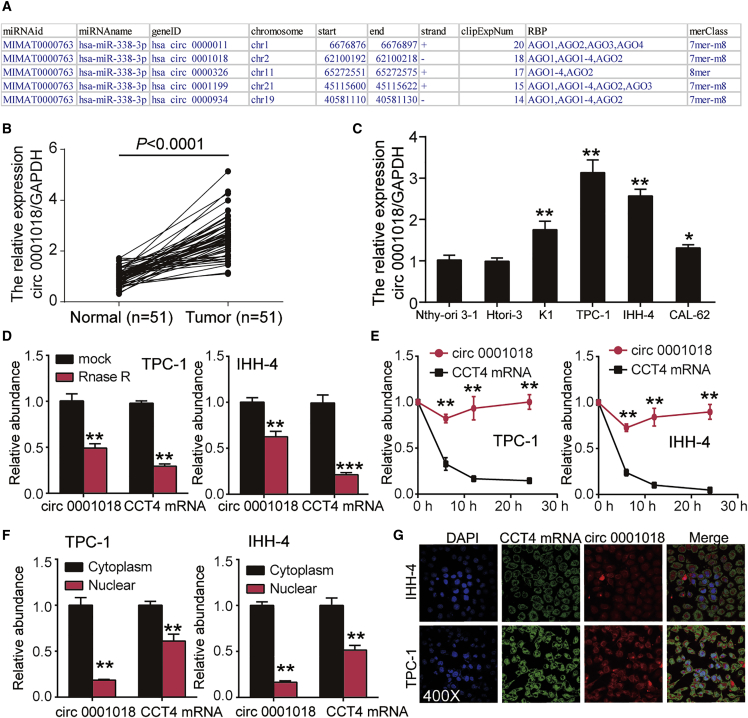
(A) An ENCORI for RNA interactomes (http://starbase.sysu.edu.cn/) algorithm was used to identify the top five most significant circular RNAs (circRNAs) based on the AGO CLIP-seq experiment support. (B) circ_0001018 levels were detected by RT-PCR in adjacent normal tissues (n = 51) compared with tumor tissue samples (n = 51); GAPDH was used as the standard reference. (C) circ_0001018 expression levels in PTC cell lines (K1, TPC-1, and IHH-4), a thyroid cell line (CAL-62), and human thyroid follicular epithelial cell lines (child Nthy-ori 3-1 cell line and parent Htori-3 cell line) were analyzed by RT-PCR; GAPDH was used as the reference. ∗p < 0.05, ∗∗p < 0.01, compared with the Nthy-ori 3-1 cell line. (D) RNase R analysis was used to determine the relative abundance of circ_0001018 and CCT4 linear mRNA in TPC-1 and IHH-4 cell lines. ∗∗p < 0.01, ∗∗∗p < 0.001, compared with the mock group. (E) Variation of relative abundance of circ_0001018 and CCT4 mRNA in TPC-1 and IHH-4 cells during a 1-day incubation. ∗∗p < 0.01, compared with linear RNA level. (F) Distribution of circ_0001018 and CCT4 mRNA in cytoplasm and nuclear fraction in TPC-1 and IHH-4 cells. ∗∗p < 0.01, compared with cytoplasm; (G) The subcellular localization of circ_0001018 and CCT4 mRNA in cells was identified using a FISH assay. Original magnification, ×400. In (C)–(F), data indicate mean ± SD from at least three independent experiments and three repetitions where cellular experiments are indicated.