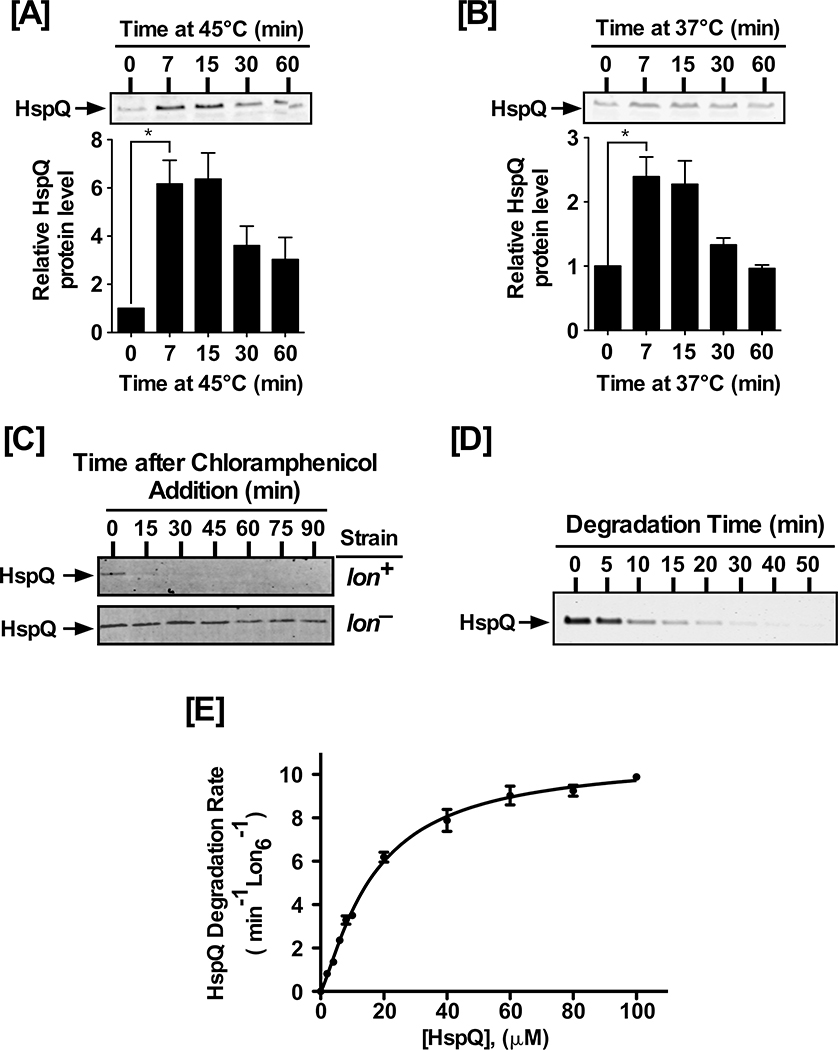
Figure 1: HspQ is a stress-induced Lon substrate.
(A) Yersinia pestis HspQ protein is induced by heat-shock at 45°C, and (B) induction of the T3SS at 37°C. Total cellular proteins, from equal number of cells at indicated time point, were examined by Western blot analysis to detect changes in HspQ protein levels, using polyclonal anti-HspQ antibodies (Key Resources Tables). p-values were calculated by performing one-tailed t-tests between HspQ protein levels at 0 min and 7 min under heat-shock (A) * p= 0.017, and T3SS induction (B) * p= 0.023. (C) In vivo degradation of HspQ requires Lon protease. lon+ and lon− Yersinia strains were cultured and chloramphenicol was added to inhibit protein synthesis. HspQ protein levels were determined by Western blot analysis. (D) In vitro proteolysis of HspQ by Lon, using 10 μM HspQ and 200 nM Lon6, was carried out in 1X Lon activity buffer in the presence of an ATP regeneration system at 37°C. Aliquots were mixed with 2X-SDS sample buffer at specified time points, resolved by electrophoresis on 15% Tris-Tricine gels, stained with Coomassie Brilliant Blue, scanned using a Li-COR Odyssey scanner, and quantified using the Image Studio software. (E) Kinetics of HspQ degradation by Lon at a range of HspQ concentrations. Degradation assays, with specified HspQ concentrations, were carried out as described in (D). Degradation rates for HspQ were fitted to a modified Hill equation. Representative Coomassie Brilliant Blue stained gels and Western blots are shown, and the data presented in graphs are from three independent experiments (mean ± SEM).