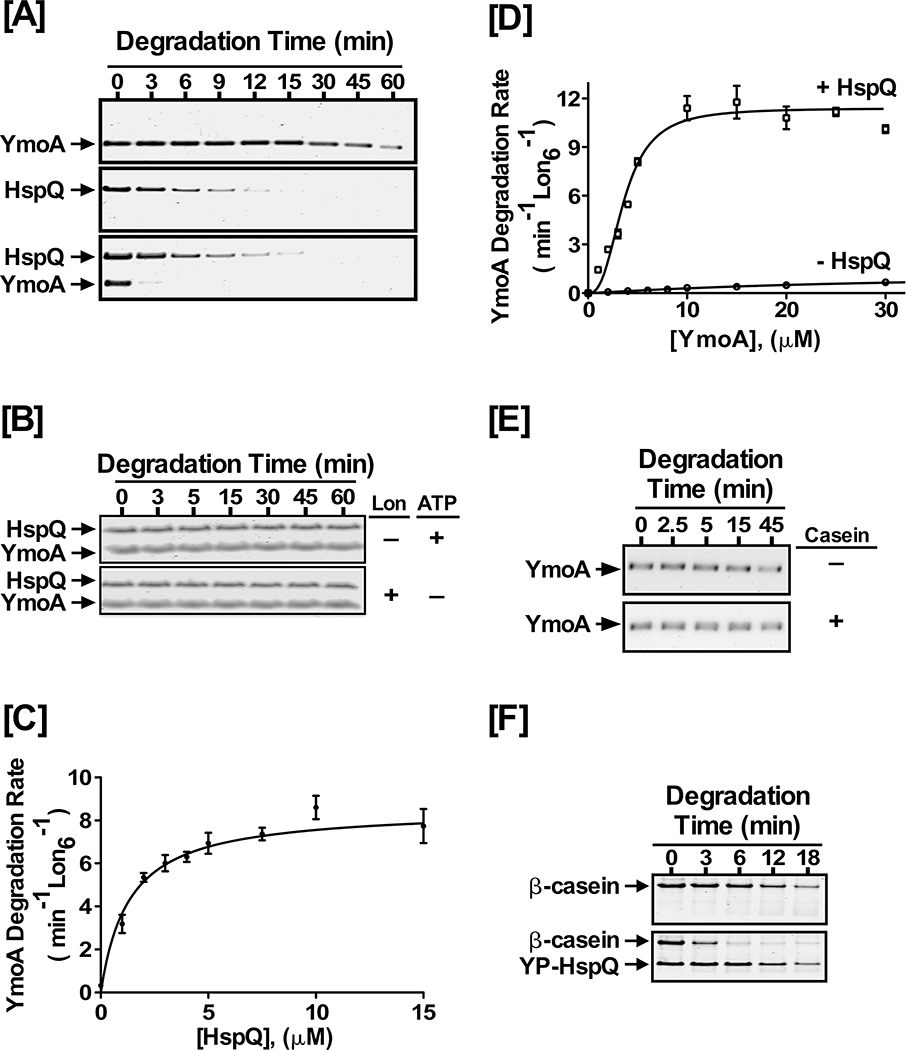
Figure 2: HspQ enhances degradation of YmoA by Lon.
Purified Yersinia YmoA protein was examined in Lon degradation assay without and with HspQ. Reactions containing equimolar concentrations of 10 μM each HspQ and YmoA were subjected to proteolysis by 200 nM Lon6 in 1X Lon activity buffer at 37°C as described in Fig 1D. (B) In vitro proteolysis assays performed in absence of Lon or ATP regeneration system are shown. Both Lon and ATP are required for HspQ-enhanced YmoA degradation. (C) The effect of HspQ on YmoA degradation by Lon was measured at a range of HspQ concentrations. Degradation rates at each HspQ concentration were measured and fitted to a Michaelis-Menton equation. (D) Kinetics of YmoA degradation by Lon. Degradation assays with a range of YmoA concentrations were carried out in absence and presence of 10 μM HspQ. Creatine Kinase (CK) is a component of the ATP regeneration system. Degradation rates at each YmoA concentration were measured and fitted to a modified Hill equation. (E) YmoA degradation by Lon was monitored in absence and presence of 100 μg/mL casein, showing that casein has no effect on YmoA degradation. (F) In vitro proteolysis of 200 μg/mL β-casein by 200 nM Lon6 was carried out in absence and presence of 10 μM HspQ. Representative Coomassie Brilliant Blue stained gels are shown.