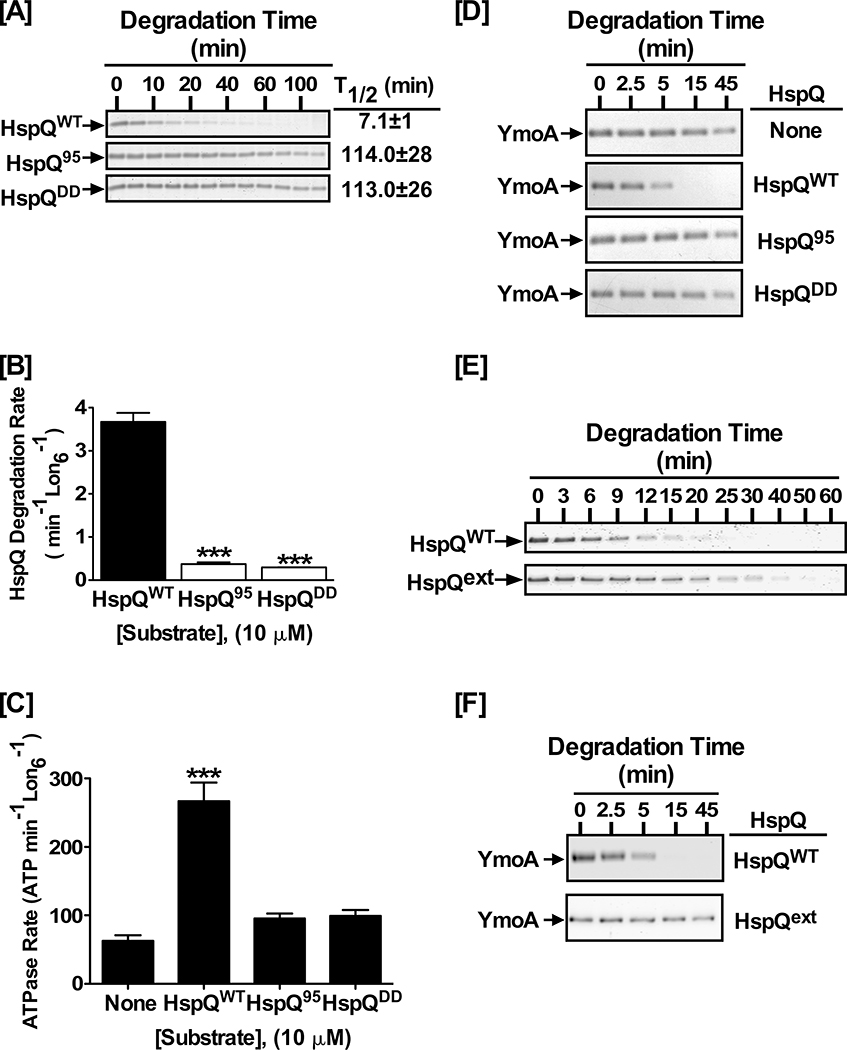
Figure 5: Precise positioning of the HspQ degron is required for Lon activation.
(A) In vitro proteolysis of 10 μM each HspQWT, HspQ95, and HspQDD by 200 nM Lon6. Reactions were carried out in 1X Lon activity buffer at 37°C. (B) Degradation rates of reactions in panel A were calculated and plotted. One-tailed t-tests were performed to calculate p-values (*** = p<0.0001). (C) HspQ degron mutants do not stimulate ATP-hydrolysis by Lon. ATP-hydrolysis assay for Lon protease in absence and presence of 10 μM HspQ C-terminal variants, as compared to wild-type HspQ, are shown with p-values (*** = p<0.0001). (D) YmoA proteolysis assays by Lon were carried out with 10 μM each HspQWT, HspQ95, and HspQDD to assess the ability of the HspQ degron mutants to allosterically activate Lon for enhanced YmoA degradation. HspQ95 and HspQDD are defective in allosterically activating Lon protease. (E) Proteolysis assays were performed with 10 μM each HspQWT and HspQext, showing that increasing the distance between the C-terminal HspQ degron and the HspQ core (HspQext) impacts its degradation. (F) In vitro Lon proteolysis of YmoA with equimolar HspQext was carried out under conditions described in (A), showing the HspQext variant does not activate Lon protease for enhanced YmoA degradation. Representative Coomassie Brilliant Blue stained gels are shown, and the data presented in graphs are from three independent experiments (mean ± SEM).