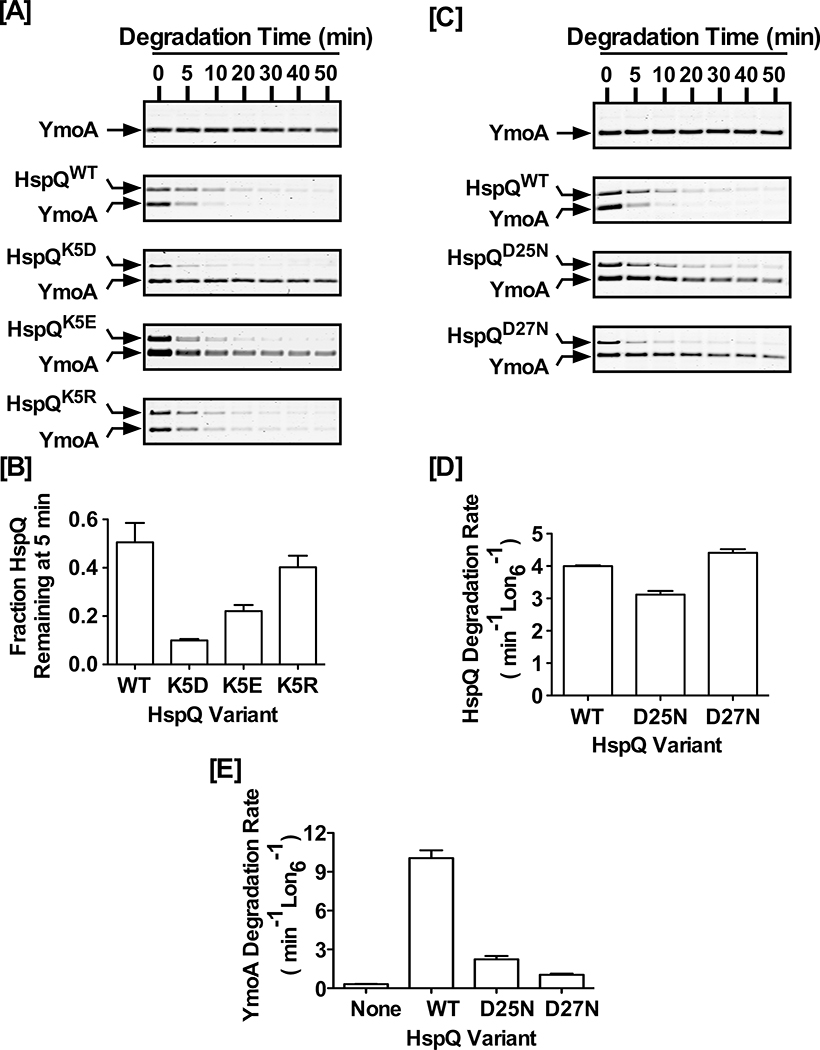
Figure 7: Conserved HspQ residues contribute to allosteric activation of Lon.
(A) The proteolytic stability of HspQWT, HspQK5D, HspQK5E, and HspQK5R and their effect on Lon activation for enhanced YmoA degradation were examined at 10 μM concentration of each HspQ variant and YmoA. Reactions were carried out in Lon activity buffer at 37°C. (B) HspQ fraction remaining at 5 min was calculated for K5 variants and plotted to illustrate the effect of side-chain length and charge on HspQ stability. (C) The proteolytic stability of HspQWT, HspQD25N, and HspQD27N and their effect on Lon activation for enhanced YmoA degradation were examined at 10 μM concentration of each HspQ variant and YmoA. (D) Degradation rates for HspQWT and its second contact site variants (HspQD25N and HspQD27N) were calculated and plotted to demonstrate that Lon degrades them at similar rates. (E) YmoA degradation rates were measured in the presence of HspQWT, HspQD25N, and HspQD27N and plotted to show that these second contact site variants are defective in allosterically activating Lon. Representative Coomassie Brilliant Blue stained gels are shown, and the data presented in graphs are from three independent experiments (mean ± SEM).