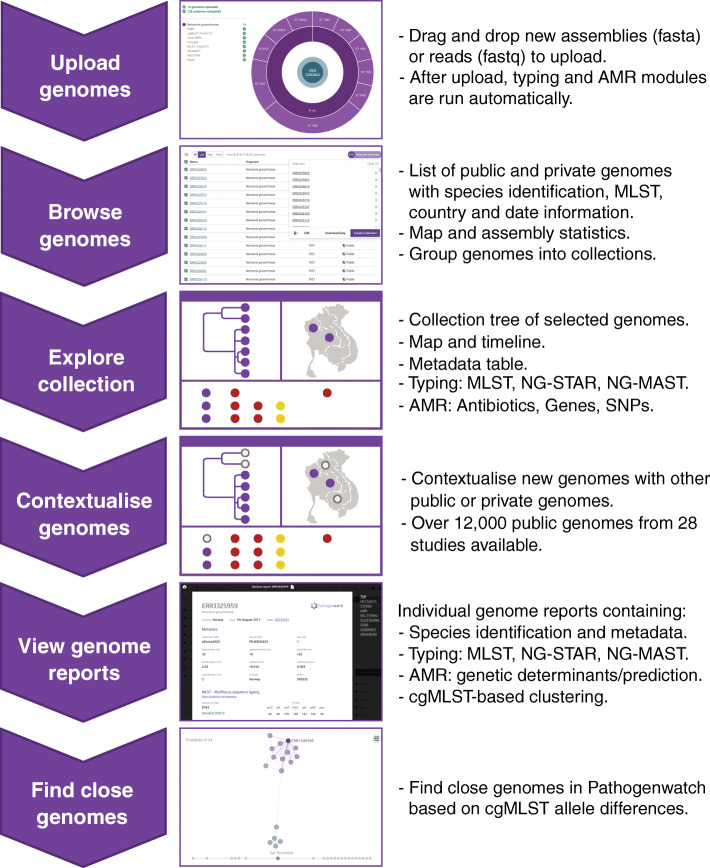
Fig. 1.
Main workflow in Pathogenwatch. New genomes can be uploaded and combined with public data for contextualisation. The collection view allows data exploration through a combined phylogenetic tree, a map, a timeline and the metadata table, which can be switched to show typing information (multi-locus sequence typing, MLST; N. gonorrhoeae sequence typing for antimicrobial resistance, NG-STAR; and N. gonorrhoeae multi-antigen sequence typing, NG-MAST) as well as known genetic AMR mechanisms for eight antibiotics. Genome reports summarise the metadata, typing and AMR marker results for individual isolates and allow finding other close genomes in Pathogenwatch based on core genome MLST (cgMLST). SNPs: single-nucleotide polymorphisms