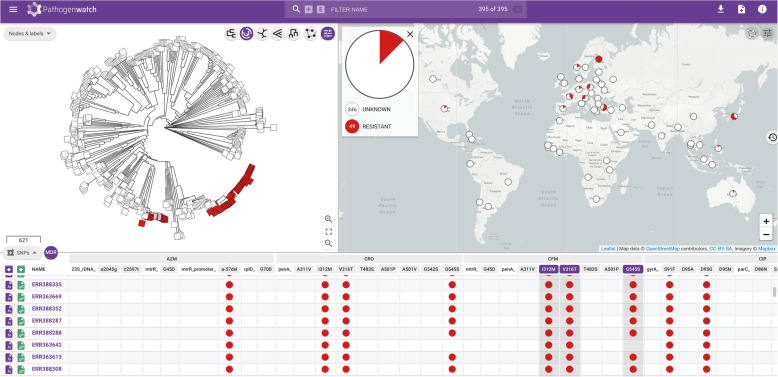
Fig. 2.
Main display of a Pathogenwatch collection, showing a phylogenetic tree, a map and a table of SNPs associated with AMR of 395 N. gonorrhoeae genomes from a global study [65, 112]. Isolates carrying three mosaic penA marker mutations are marked in red in the tree and the map. The table can be switched to show the metadata, a timeline, typing results (multi-locus sequence typing, MLST; N. gonorrhoeae sequence typing for antimicrobial resistance, NG-STAR and N. gonorrhoeae multi-antigen sequence typing, NG-MAST) and AMR analytics (known genetic mechanisms and genotypic AMR prediction) implemented in the platform. Further detail is shown in Additional file 3: Fig. S1. The contents of and boundaries in the map are the sole responsibility of Pathogenwatch and do not necessarily reflect the views or opinions of WHO or other Public Health Agency