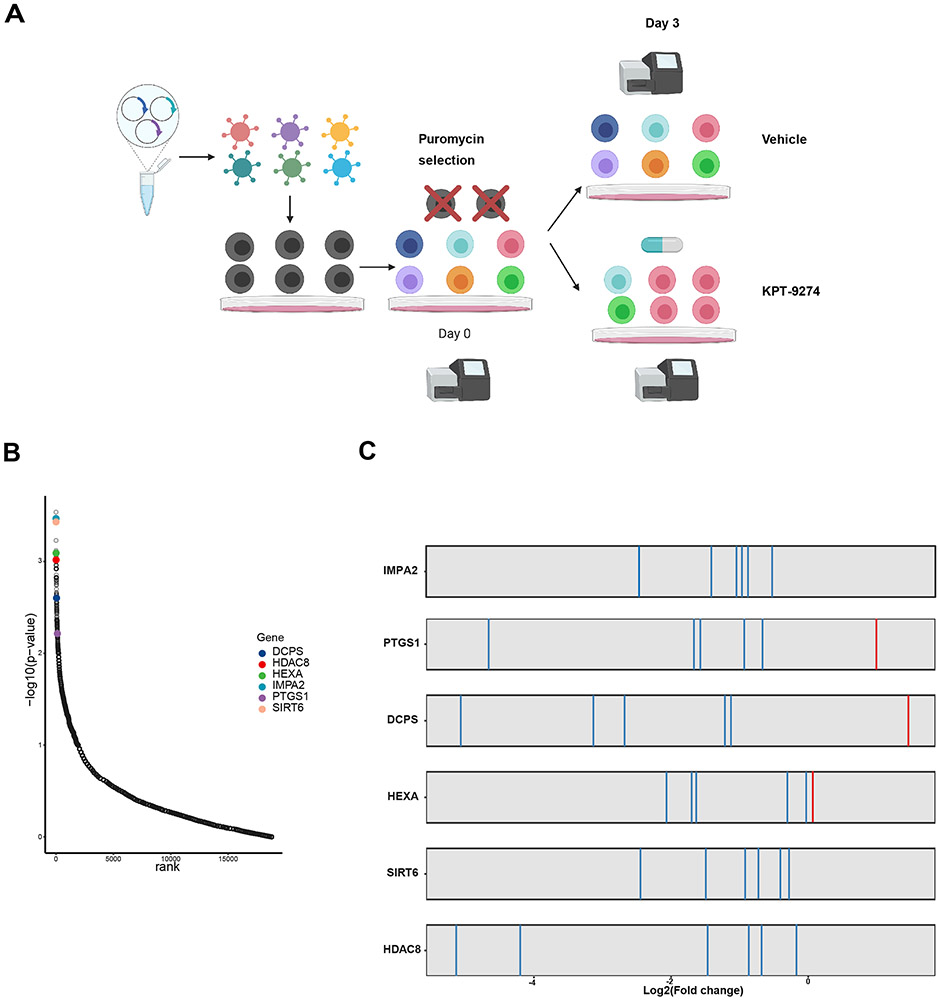
Figure 1. Genome-wide functional screening identifies two histone deacetylase genes as combination targets for NAMPTi.
A) Schematic illustration of CRISPR screen workflow. CRISPR screen was conducted by transfecting GeCKOv2 library into MOLM13 cells. Virus MOI was titrated to ensure a single sgRNA to be transduced into each cell. 50 nM KPT-9274 or DMSO vehicle was administrated to CRISPR library-transfected MOLM13 cells after puromycin selection. B) Top scored genes ranked by their −log(p-values) and displayed against each gene’s corresponding ranks of essentiality. Top hits for validation are highlighted. p-values of genes were calculated based on sgRNA efficiency. C) sgRNAs targeting selected top hits are constantly depleted following KPT-9274 treatment.