Figure 4: (N)RAS melanomas frequently experience clonal mutations in the PBAF complex, and PBAF complex mutations are associated with improved overall survival (OS) and progression free survival (PFS) when treated with immunotherapy.
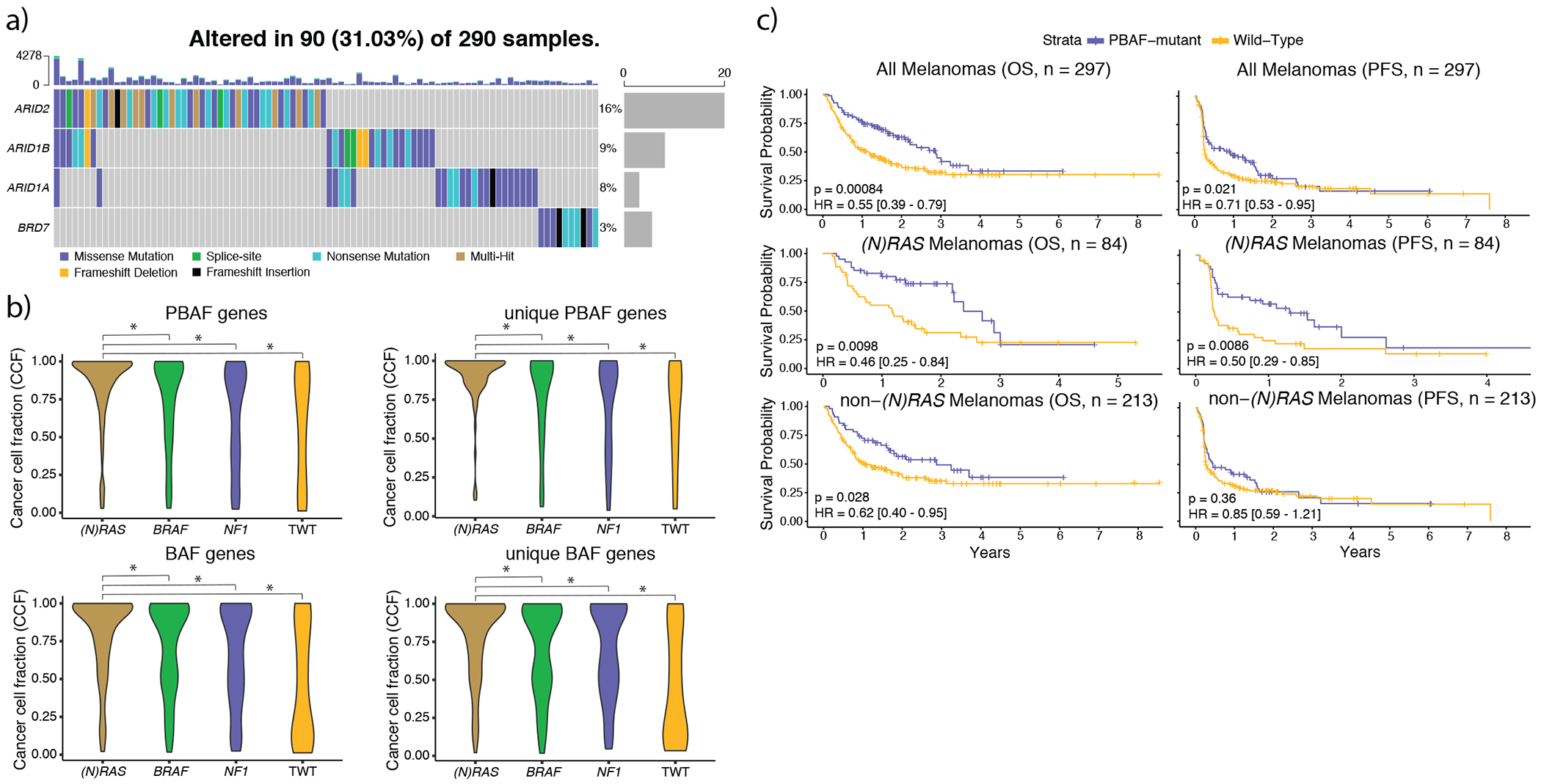
a) The co-mutation plot of BAF/PBAF complex SMGs identified in (N)RAS melanomas. Putative loss-of-function mutations (nonsense, splice-site, indels) are almost entirely mutually exclusive with one another. Further, mutations in ARID2 and BRD7 (specific to the PBAF version of the SWI/SNF complex) were never observed in the same tumor. b) The distributions of cancer cell fractions (Methods) for all PBAF and BAF complex genes among the genomic subtypes. Mutations in BAF/PBAF complex genes were enriched for being clonal (Methods) in (N)RAS melanomas compared to other genomic subtypes (χ2 pairwise adjusted for subtype proportions, p < 2.24 x 10−4, two-sided), and PBAF gene mutations were clonal more frequently than BAF gene mutations in (N)RAS melanomas (p = 0.003, Kolmogorov-Smirnov, two-sided). An asterisk denotes a Kolmogorov-Smirnov p-value < 0.05. c) Mutations in PBAF genes are associated with significantly improved OS and PFS to immunotherapy. Although PBAF-mutant (N)RAS and non-(N)RAS melanomas have significantly better OS compared to their PBAF wild-type counterparts, the improvement in OS is much more pronounced in (N)RAS melanomas. PBAF-mutant non-(N)RAS melanomas do not experience significantly better PFS compared to PBAF wild-type non-(N)RAS melanomas. However, PBAF-mutant (N)RAS melanomas have significantly improved PFS compared to PBAF wild-type (N)RAS melanomas, and the PFS signal from PBAF-mutant (N)RAS melanomas are driving the significant improvement in PFS at the entire cohort level.
