Figure 5: Identification of novel drivers and enrichment of mutational signature 3 in triple wild type (TWT) melanomas.
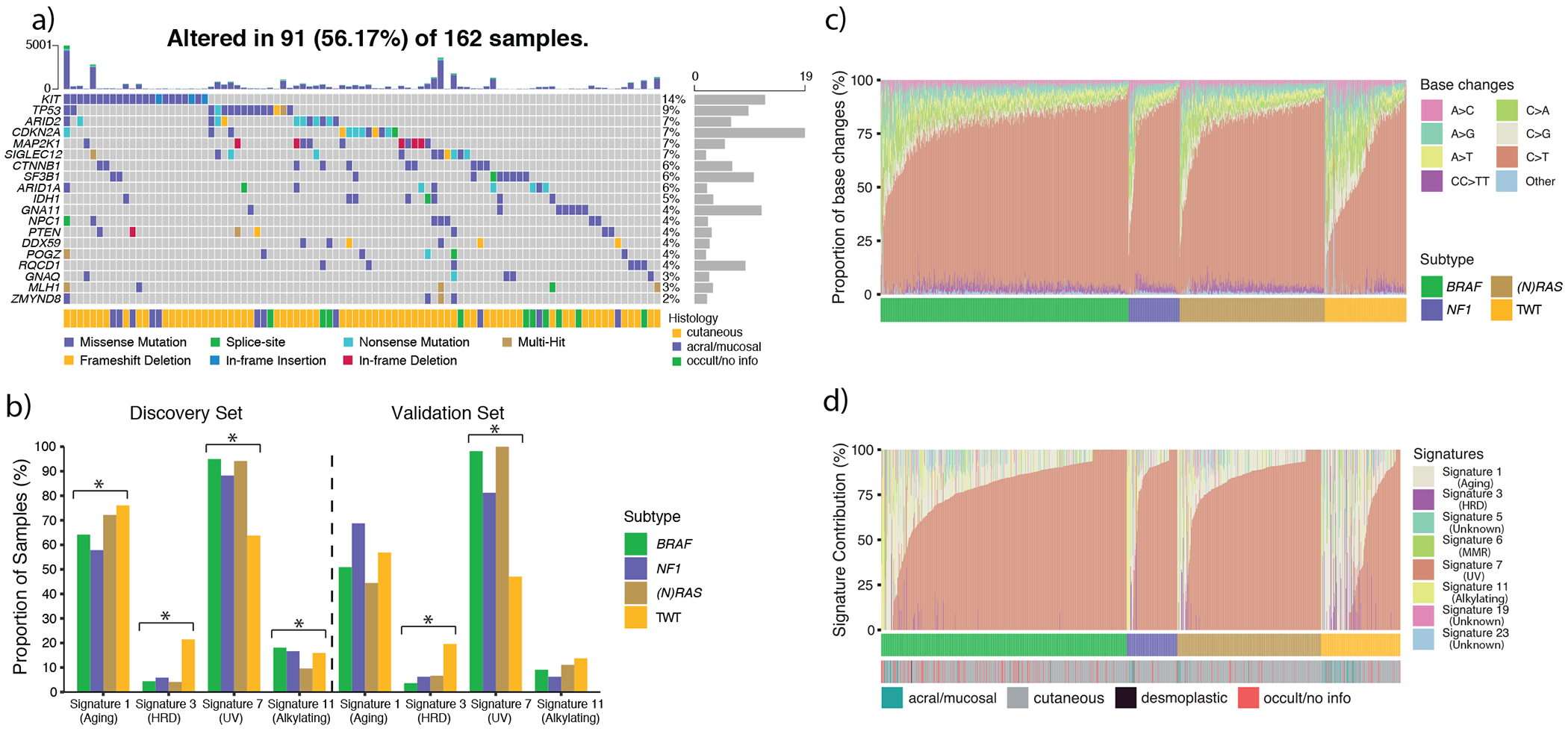
a) The co-mutation plot of TWT significantly mutated genes (SMGs), including the annotation of tumor histology. These SMGs include canonical melanoma cancer genes (e.g. CTNNB1, CDKN2A, TP53) and known uveal melanoma driver genes (e.g. GNA11, GNAQ, and SF3B1). However, these 19 SMGs only explain the presence of drivers in just over 50% of TWT melanomas. b) The proportion of samples in each genomic subtype exhibiting mutational signatures (Methods) in our discovery and validation (n = 159) cohorts. Signature 3 was present in 21.5% of TWT melanomas compared to 4.5% of non-TWT melanomas (p = 2.20 x 10−11, Fisher’s exact test, two-sided), and was the third most active mutational signature in TWT melanomas. In our validation cohort (Methods), signature 3 was identified in 19.6% of TWT melanomas and 5.6% of non-TWT melanomas (p = 0.001, Fisher’s exact test, two-sided), and was again the third most active signature in TWT melanomas. In both the discovery and validation cohorts, the proportion of tumors with signatures 3 and 7 were significantly different across the genomic subtypes (p < 0.05, χ2, two-sided). An asterisk denotes a χ2 p-value < 0.05. c) The proportion of base changes ordered by genomic subtype and the proportion of C>T transitions (increasing, left-to-right). d) The relative contribution of each mutational signature ordered by genomic subtype and the relative signature 7 contribution (increasing, left-to-right).
