Extended Data Fig. 3. PBAF complex immunotherapy validation (overall survival and RECIST response).
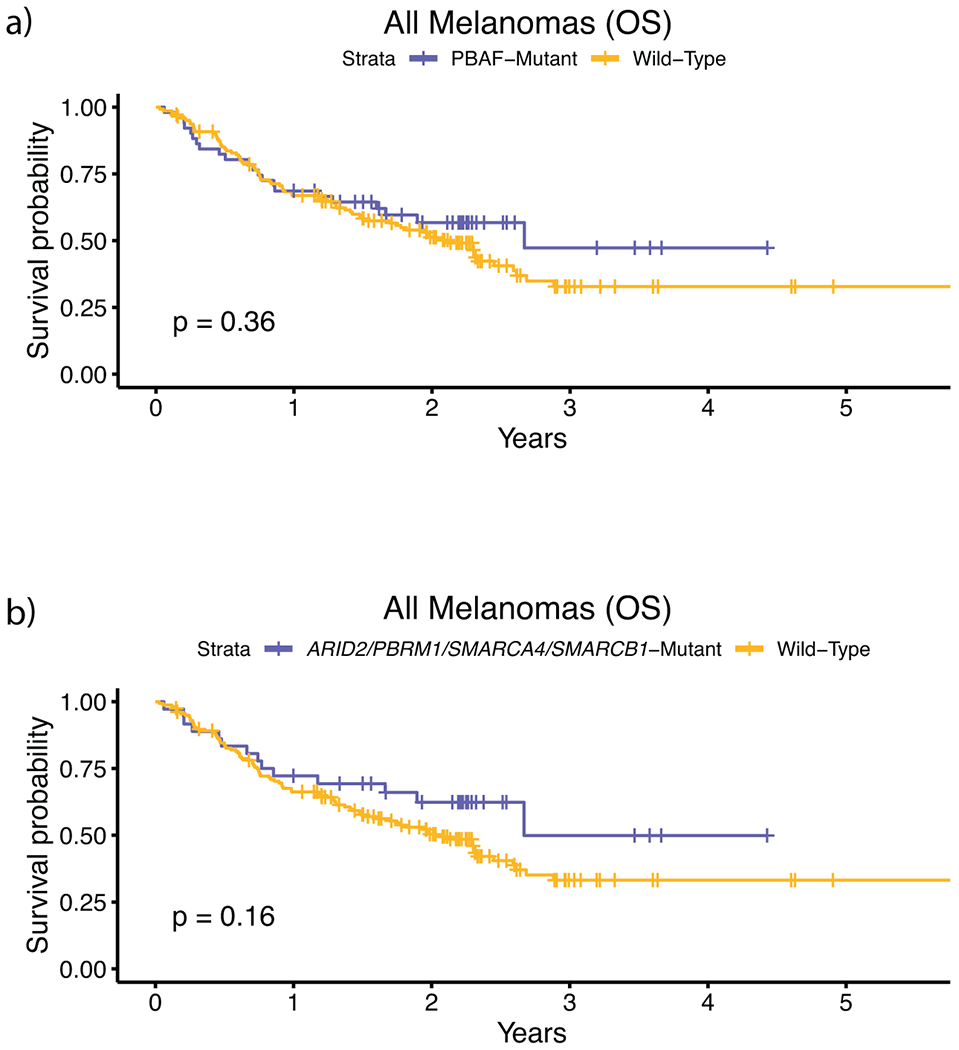
External validation analysis of overall survival for PBAF mutations using the Roh, Riaz, Hugo, and Rodig cohorts (n = 194), which are immunotherapy treated, whole-exome sequenced, cohorts not included in our discovery cohort. a, Survival curves between PBAF-mutants and non-PBAF mutants. b, Survival curves between PBAF-mutants and non-PBAF mutants where PBAF mutants are classified by having mutations in ARID2, PBRM1, SMARCA4, and SMARCB1, which are the 4 PBAF complex genes commonly used in clinical sequencing panels. This limited validation cohort lacked sufficient samples with co-mutation of (N)RAS and PBAF complex genes (n = 9), and thus validation analysis was only performed on all tumors. Due to the unique treatment regimens in each of these cohorts, we were unable to correct for drug. Further, because we did not have access to raw sequencing data from these studies, we could not calculate and correct for tumor purity. When correcting only for mutational load the hazard ratio of PBAF mutations in the whole-exome cohorts, (a) when considering all genes in the PBAF complex, was 1.07 (multivariate Cox proportional-hazards, p = 0.80). The differences in these findings relative to the primary larger cohort may indicate differences in patient population and study size relative to our discovery cohort. (b) When considering only mutations in ARID2, PBRM1, SMARCA4, and SMARCB1 as PBAF-mutant, the HRR was 0.86 (multivariate Cox proportional-hazards, p = 0.61). The p-values for a-b) are derived from the log-rank test.
