Figure 2.
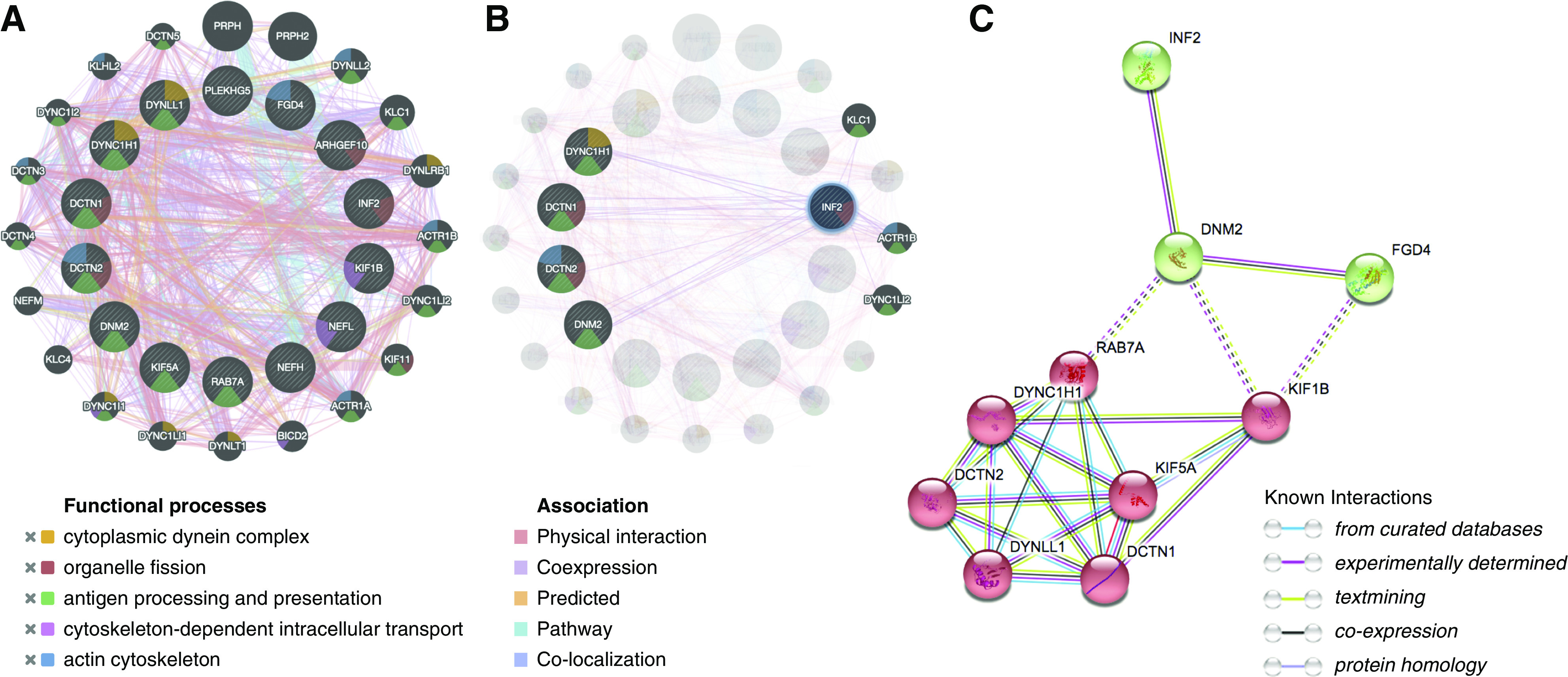
Bioinformatic analysis suggests INF2 participates in the dynein trafficking pathway. (A and B) PPI generated in GeneMANIA (13 CMT-causing genes ARHGEF10, DCTN1, DCTN2, DNM2, DYNC1H1, FGD4, INF2, KIF1B, KIF5A, NEFH, NEFL, PLEKHG5, RAB7A, and Dynll1 are drawn in the inner cycle, whereas 20 predicted genes by the molecular function–based weighting method are located in the outer circle. Distinct colors of the network edge indicate the bioinformatics methods applied: physical interactions, coexpression, predicted, colocalization, and pathway. The different colors used for the nodes indicate the biologic functions of the sets of enrichment genes: cytoplasmic dynein (orange), cytoskeletal-based intracellular transport (purple), organelle fission (red), and surface protein processing and presenting (green). (C) STRING generated a PPI with significant enrichment (PPI enrichment P<0.001). The network was further categorized into two subnetworks (in red and in green) by K-means clustering. The intercluster edges are represented by dashed lines.
