Figure 6.
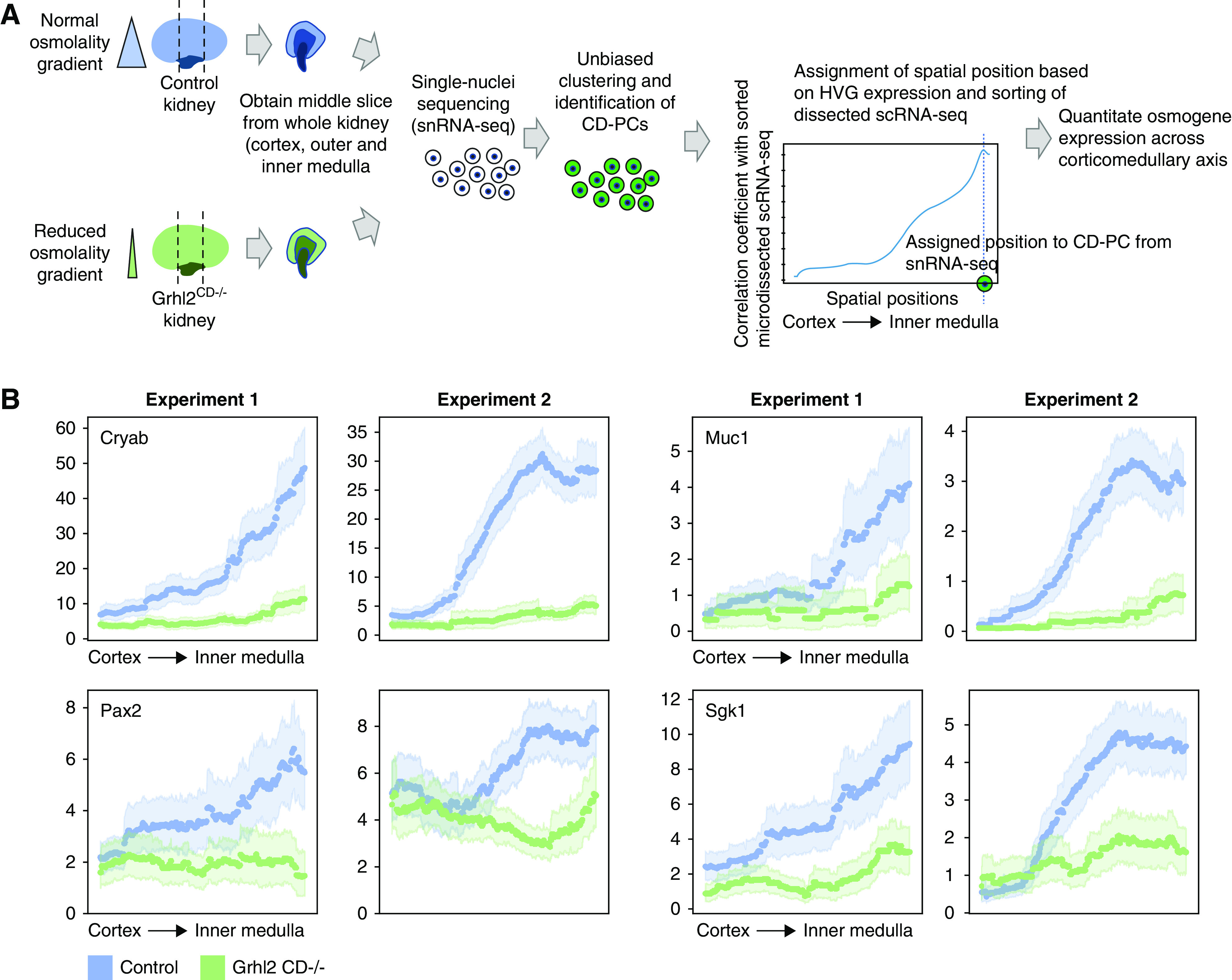
In silico spatial sorting enables physiologic phenotyping of the renal osmolality gradient in whole kidney samples. (A) Spatial sorting of CD-PCs from whole kidney snRNA-seq data. Depicted is the experimental strategy to assign spatial positions to CD-PCs in whole kidney snRNA-seq data. We chose to perform snRNA-seq to overcome the underrepresentation of medullary cells in whole kidney scRNA-seq. To further enrich for medullary cells, we obtained a middle slice containing the cortex, outer medulla, and inner medulla from each kidney of Grhl2CD−/− mice (decreased intrarenal osmolality gradient) and control mice (normal intrarenal osmolality gradient). Single-nuclei transcriptomes were obtained, and CD-PCs were identified on the basis of unbiased clustering and marker gene expression. CD-PCs from snRNA-seq were assigned regional positions by correlating HVG expression of each nucleus with HVG expression along the corticomedullary axis of sorted dissected samples (sorting performed with 500 positions, Figure 3B and C). We assigned the spatial position with maximum correlation as the new spatial position of the respective nucleus. This enabled the analysis of osmogene expression along the corticomedullary axis in the whole kidney snRNA-seq data. (B) Regional expression of selected osmogenes on the basis of snRNA-seq correctly predicts the reduced intrarenal osmolality gradient in Grhl2CD−/− mice. Depicted is a sliding average over 150 spatial positions for each whole kidney snRNA-seq sample (dotted lines with 95% confidence intervals indicated by light shades). Results are shown for two independent biologic replicates (experiment 1 and experiment 2). Expression values are library-normalized raw counts. Decreased osmogene expression in the medulla of Grhl2CD−/− kidneys reflects the known reduced medullary osmolality in these mice.19
