Figure S5.
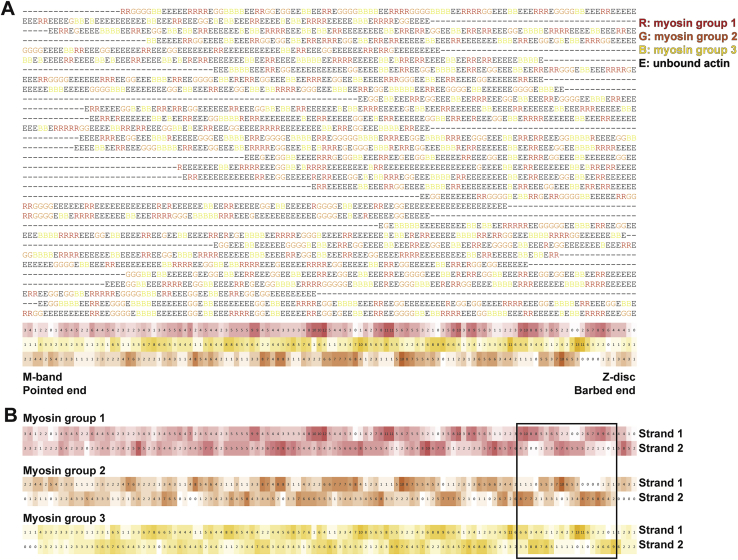
Overall myosin binding profile generated from multiple sequence alignment of myosin binding profiles of 30 thin filaments, related to Figure 4 and STAR Methods
(A) The fitted myosin head model in Figure 3B was converted to myosin binding profile sequences on thin filaments, with “R,” “G,” “B” representing actin subunits bound by three groups of myosin heads originating from three neighboring thick filaments and “E” representing myosin-free actin subunits. These sequences were aligned using multiple sequence alignment with a customized weight matrix. Only the sequence of one actin strand for each thin filament was used for alignment. The sum of occurrence of each myosin group at each actin subunit position is shown at the bottom and colored accordingly. Darker color indicates higher occurrence.
(B) The colored overall myosin binding profile on both actin strands for each myosin group. Sequences of strand 2 were combined using the alignment of strand 1 shown in (A). The overall myosin binding profiles depict hotspots for myosin binding on a thin filament and the region highlighted in the black box is used to color the models shown in Figure 4B.