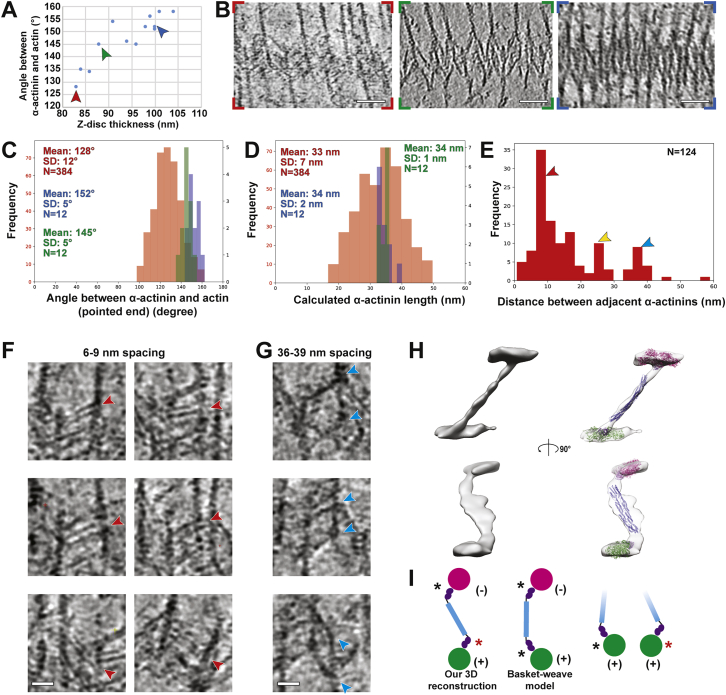
Figure S7.
α-Actinin structure and organization in the Z-disc, related to Figure 7
(A) Plot relating the thickness of Z-discs and the angle between α-actinins and the pointed end of actin filament from different tomograms. The positive correlation implies a parallel hinge mechanism of the Z-disc.
(B) Slices through tomograms depicting a thin Z-disc (left, red arrow in (A)), a thick Z-disc (right, blue arrow in (A)) and a Z-disc of intermediate thickness (middle, green arrow in (A)). Scale bar, 50 nm.
(C) Distribution of the calculated angles between annotated α-actinins and actin filaments in direction to the pointed end in the thin (red), thick (blue) and intermediate-thickness (green) Z-discs. The y axis for both the thick and intermediate Z-disc is shown on the right in black. There are more data points for the thin Z-disc as it was completely annotated, while a few examples of α-actinin were selected for the intermediate and thick Z-discs.
(D) Distribution of the length of annotated α-actinins in the thin (red), thick (blue) and intermediate-thickness (green) Z-discs. The distance along α-actinin between the centers of the connecting actin filaments was measured and the length of α-actinin was calculated by subtracting the diameter of an actin filament (6 nm). The relatively large standard deviation in the thin form Z-disc is likely caused by α-actinins binding to actin filaments at different azimuthal orientations and the error in the precise determination of the central axis of actin filaments.
(E) Distribution of the calculated spacing between adjacent α-actinins in the thin Z-disc. The red, yellow, and blue arrow heads highlight peaks at 6-9 nm, 24-27 nm and 36-39 nm, respectively. The 24-27 nm peak appears as a result of the two other types of spacing (36 - 2x6).
(F and G) Example images showing α-actinins with the 6-9 nm spacing (D) and the 36-39 nm spacing (E). Arrow heads highlight the positions and orientations of α-actinins. Scale bar, 20 nm.
(H) 3D reconstruction of α-actinin obtained from sub-volume averaging. Although there is a strong missing wedge effect resulting in an elongation of the reconstruction in one direction, we were able to manually fit atomic models derived from the crystal structure of α-actinin (PDB: 4D1E) and the cryo-EM structure of actin filaments bound by the first calponin homology domain of the actin binding domain (PDB: 6D8C) into the density. Actin filaments are shown in magenta and green; the actin binding domain and the rod region are depicted in purple and blue, respectively. The flexible neck regions and the C-terminal calmodulin-like domains are not shown.
(I) Left: Schematic diagram showing the difference between averaged α-actinin structure and the conventional basket-weave model. Right: The two different interactions (marked by the red and black asterisks) between the end of α-actinin and actin filaments (magenta and green) are demonstrated on the right, formed by the flexible neck region between the rod (blue) and actin binding domain (purple).