Figure 1. KEGG2Net workflow and directed acyclic graph (DAG) generation.
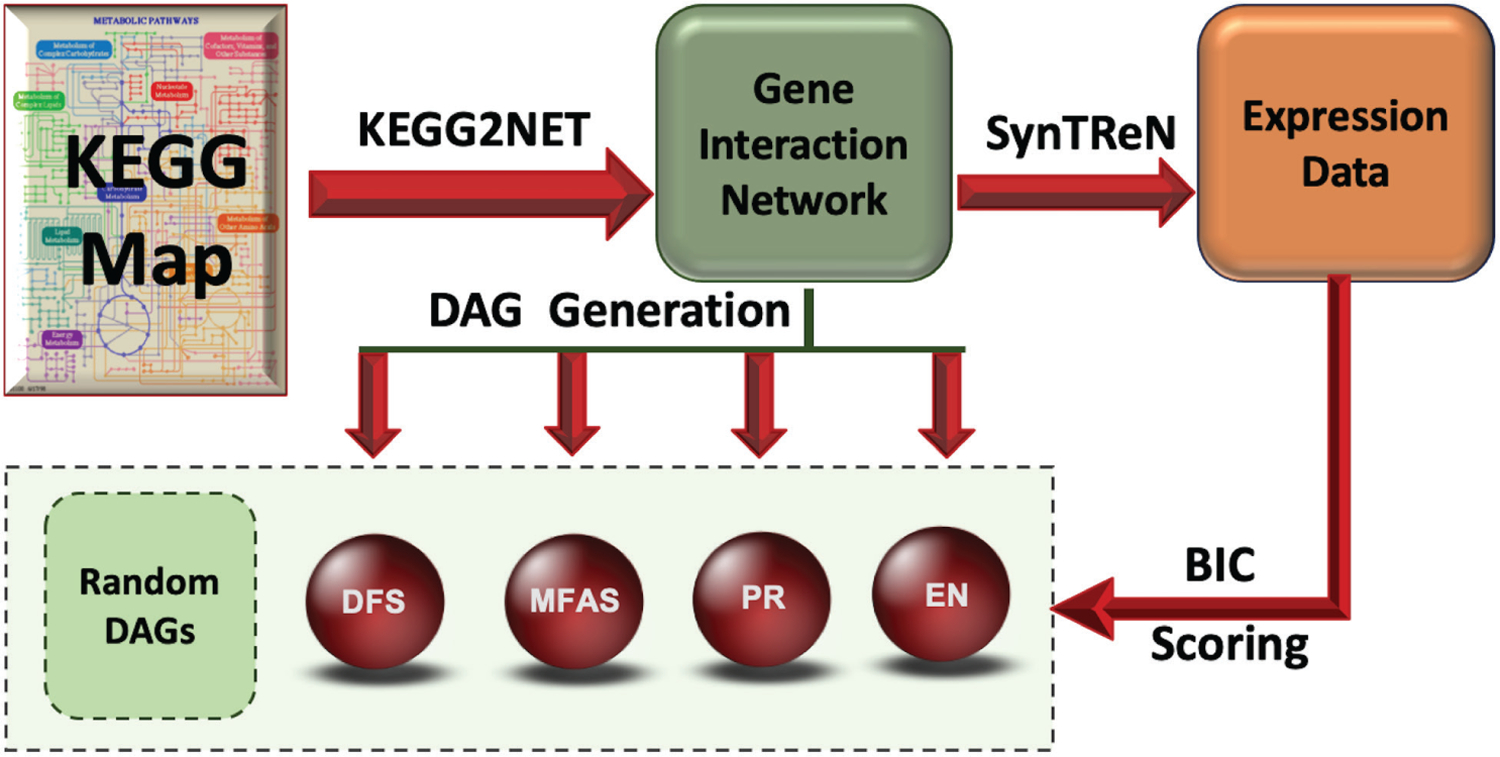
Each KEGG pathway is converted into a gene interaction network where only the genes in the pathway are represented, and all direct and indirect interactions among the genes are preserved. For each network, four DAGs (using four alternative cycle removal methods depth-first search (DFS), minimum feedback arc set (MFAS), PageRank (PR), and ensemble (EN)) and 1,000 random DAGs for each of the four DAGs are generated. The random DAGs follow the node and edge statistics of their corresponding DAG. For each gene interaction network, synthetic gene expression data is generated using SynTRen and the fitness of the four DAGs (and the corresponding 4×1,000 = 4,000 random DAGs) is assessed using Bayesian Information Criterion (BIC) scoring.
