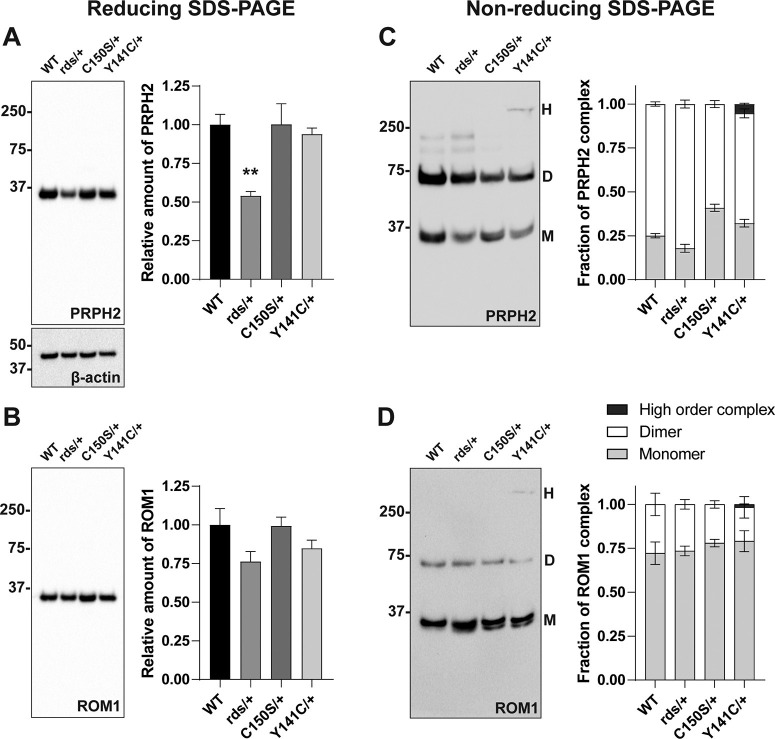
Figure 2.
The C150S and Y141C heterozygous mutations affect PRPH2 supramolecular organization without causing a significant reduction in the PRPH2 expression level. A, B, A single Western blot probed for both PRPH2 and ROM1 after protein separation by SDS-PAGE under reducing conditions. Each sample contained 10 µg of lysate obtained from eyecups of WT, rds/+, C150S/+, or Y141C/+ collected at P16. The amount of PRPH2 and ROM1 was quantified on the same blot using densitometry of at least four independent samples, normalized to β-actin as a loading control and plotted in the graph on the right relative to WT. One-way ANOVA with Dunnett's multiple comparisons post hoc test revealed a statistically significant reduction in PRPH2 for rds/+ (**p = 0.0096) but not C150S/+ or Y141C/+ eyecups (p = 0.9999 and p = 0.9346, respectively), while there was no statistical significance in ROM1 levels. C, D, A single Western blot probed for PRPH2 and ROM1 performed after protein separation by SDS-PAGE under nonreducing conditions. Each sample contained 10 µg of lysate obtained from eyecups of WT, rds/+, C150S/+, or Y141C/+ collected at P16. Under these conditions, PRPH2 and ROM1 run as abnormal high order complexes (H, >250 kDa), dimers (D, ∼75 kDa), and monomers (M, ∼37 kDa). Protein amount in each band was quantified using densitometry of three independent samples and plotted in the graph on the right. Two-way ANOVA with Dunnett's multiple comparisons post hoc test revealed that more PRPH2 exists as a monomer than dimer in C150S/+ eyecups (monomer, p < 0.0001; dimer, p < 0.0001) as well as smaller changes in both rds/+ (monomer, p = 0.0243; dimer, p = 0.0243) and Y141C/+ eyecups (monomer, p < 0.0001, dimer, p = 0.0002), while there was no statistical significance in any differences in ROM1 complexes. The high order complex that includes both PRPH2 and ROM1 was only observed in Y141C/+ eyecups.