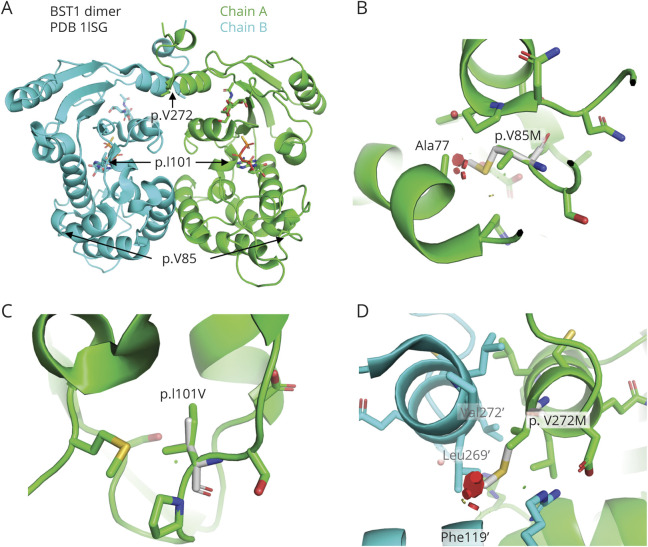
Figure. Structural Analysis of Human BST1 Variants.
This figure was produced using PyMol software version 2.2.0 and represents the following. (A) Structure of the BST1 dimer bound to ATP-γS (pdb 1ISG). Position of each variant sites is indicated. ATP-γS molecule in the active site is shown as sticks. (B) Close-up view of the p.V85M variant site. Mutated residue is shown in white. Variant would create clashes (red disks) with nearby Ala77 in the core. (C) Close-up view of the p.I101V variant site. Residue is located in the core of the protein, but the variant to a smaller residue results in no clash. (D) Close-up view of the p.V272M variant site. Residues with a prime correspond to chain B. This residue is located at the dimer interface, and the variant would create clashes with the other chain, resulting in a destabilization of the dimer.