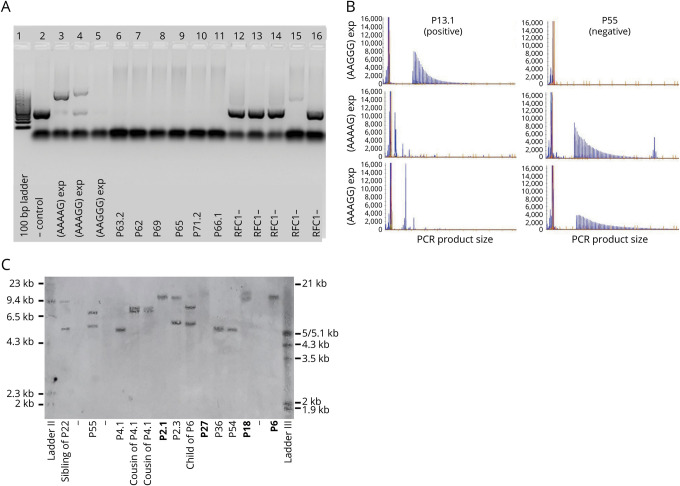
Figure 1. Identification of RFC1 Repeat Expansions.
(A) Flanking PCR, products run on a 1% agarose gel. Lanes 2 to 5: controls, including reference sequence (AAAAG)11 in lane 2. Lanes 6 to 11: positive samples with absent product amplification indicating biallelic pathogenic AAGGG expansions. Lanes 12 to 16: negative samples with product amplification consistent with reference sequence or a nonpathogenic expansion. (B) Repeat-primed PCR targeting the pathogenic (AAGGG)exp, nonpathogenic (AAAAG)exp, and (AAAGG)exp expansions; visualization of separated fluorescein amidite–labeled PCR products. Ladder markers are 35, 50, 75, 100, 139, 150, 160, 200, 250, 300, 340, 350, 400, 450, 490, and 500 nucleotides. Representative plots from a P13.1 carrying the biallelic AAGGG repeat expansion and P55 carrying nonpathogenic (AAAGG)exp/(AAAAG)exp expansions in compound heterozygous state. (C) Southern blotting of genomic DNA from 13 patients and unaffected relatives. Patients carrying biallelic pathogenic expansions in RFC1 (bold) show 2 discrete or 1 overlapping band. In RFC1-negative cases, either one 5-kb band corresponding to the expected size for reference allele (AAAAG)11 or bands of increased size can be observed.