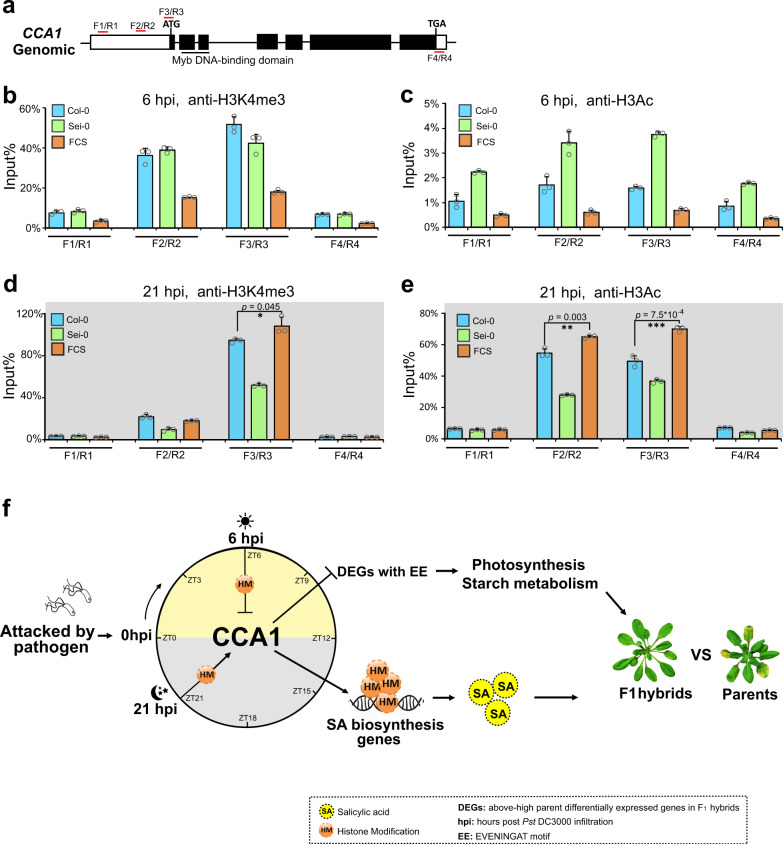
Fig. 5. Expression variations of CCA1 in hybrids correlated with its altered histone modification rhythms.
a Regions of CCA1 used for the ChIP-qPCR assays. ChIP-qPCR analyses of four fragments (shown at a) of CCA1 in the wild-type F1 hybrids and parents using an anti-H3K4me3 antibody at 6 hpi (b) and 21 hpi (d). ChIP-qPCR analyses of four fragments (shown at a) of CCA1 in the wild-type F1 hybrids and parents using an anti-H3Ac antibody at 6 hpi (c) and 21 hpi (e). ChIP values in b–e were normalized to their respective DNA inputs. The results are representative of three biological replicates, with measurements taken from independent samples grown and processed at different times. Data are shown as the mean ± SD (n = 3, n indicates biological replicates). f Working model of how CCA1 coordinates enhanced heterosis for defense and for biomass in hybrids under pathogen invasion. *p value < 0.05; **p value < 0.01; and ***p value < 0.001 (two-tailed Student’s t test).