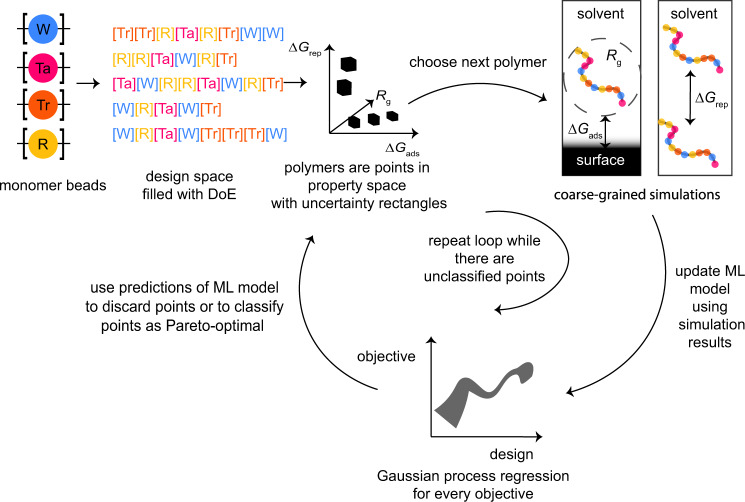
Fig. 1. Overview of the workflow.
Using classical design of experiments (DoE), we enumerate representative samples in the design space of monomer sequences, which we then explore in the active learning loop with the ϵ-PAL algorithm. For this algorithm, Gaussian process surrogate models provide us with predicted means and standard deviations (SDs) that enable us to decide which designs we can confidently discard, classify as Pareto optimal, and determine which simulation we should run next to maximally reduce the uncertainty for points near the Pareto front. Models that are trained over the course of this process can reveal structure–property relationships and can be inverted using genetic algorithms to further explore the design space.