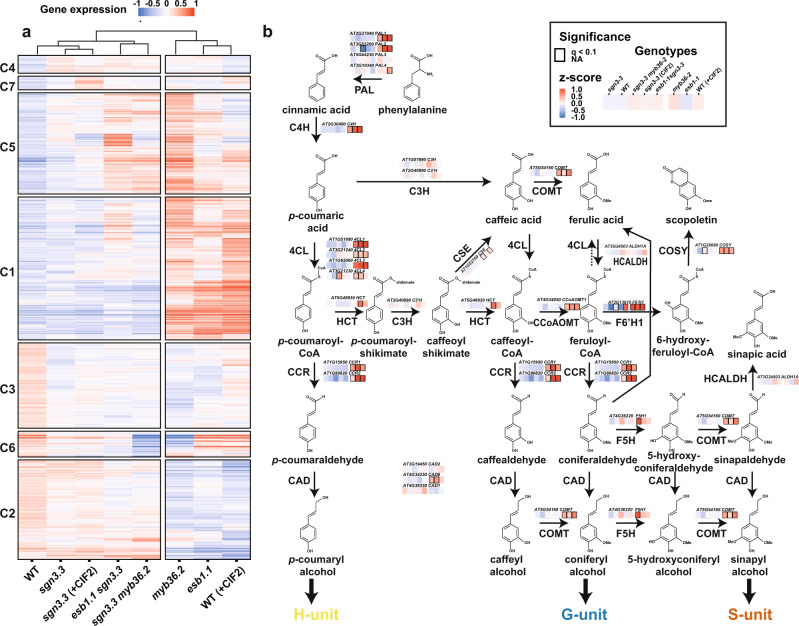
Fig. 3. Modulation of the phenylpropanoid pathway by the Schengen-pathway.
a Heatmap of the 3564 differentially expressed genes identified in the RNA-seq in root tips of wild-type (WT), sgn3-3, esb1-1, myb36-2, esb1-1 sgn3-3, sgn3-3 myb36-2 plants. Treatment with 100 nM CIF2 was applied as indicated (+CIF2) for WT and sgn3-3 plants. Clusters (C) are designated with numbers (n = 7). Genes belonging to each cluster are listed in Supplementary Data 1. b Phenylpropanoid pathway leading to the lignin monomers and scopoletin biosynthesis (adapted from72). Solid arrows represent enzymatic steps. Gene expression from the genes selected in Supplementary Fig. 3d was mapped on the pathway according to their KEGG enzyme nomenclature. Only the genes with a demonstrated function in lignin biosynthesis as listed in Supplementary Fig. 3d were mapped. PAL PHENYLALANINE AMMONIA-LYASE, C4H CINNAMATE 4-HYDROXYLASE, 4CL 4-COUMARATE:CoA LIGASE, HCT p-HYDROXYCINNAMOYL-CoA:QUINATE/SHIKIMATE p-HYDROXYCINNAMOYLTRANSFERASE, C′3H p-COUMARATE 3′-HYDROXYLASE, C3H COUMARATE 3-HYDROXYLASE, CSE CAFFEOYL SHIKIMATE ESTERASE, CCoAOMT CAFFEOYL-CoA O-METHYLTRANSFERASE, CCR CINNAMOYL-CoA REDUCTASE, F5H FERULATE 5-HYDROXYLASE, COMT CAFFEIC ACID O-METHYLTRANSFERASE, CAD CINNAMYL ALCOHOL DEHYDROGENASE, HCALDH HYDROXYCINNAMALDEHYDE DEHYDROGENASE, COSY COUMARIN SYNTHASE, F6′H1 FERULOYL COA ORTHO-HYDROXYLASE 1.