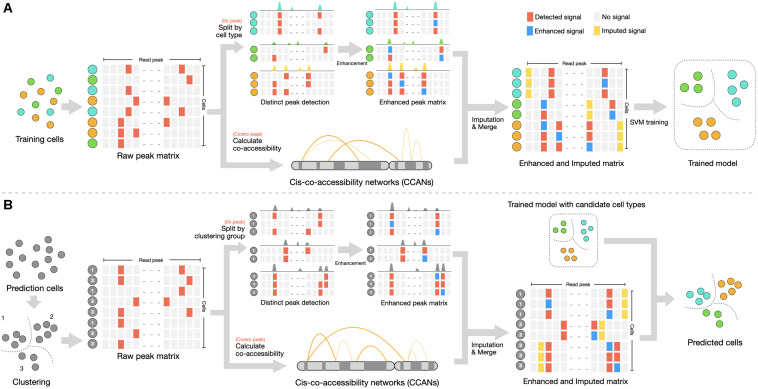
FIGURE 1.
Summary of svmATAC method. (A) Training step. A fixed-size cell-peak matrix is constructed from a labeled scATAC-seq dataset. The peak matrix will be filtered and (1) matrix with 5 k peak will be split by cell types for calling distinct peak and signal in qualified peaks will be enhanced; (2) matrix with Cicero peak will be input to Cicero for calculating co-accessibility score between peaks. Two peaks with cis-regulatory interaction will be integrated for imputation in each enhanced peak matrix. All enhanced and imputed peak matrix will be merged to a single matrix for training SVM classifier. (B) Prediction step. A clustering is necessary at first for assigning each cell a clustering group number and then the matrix will be filtered, followed by enhancement and imputation steps. Finally, an SVM classifier will identify the cell types of predicting dataset using the SVM model trained from step A.