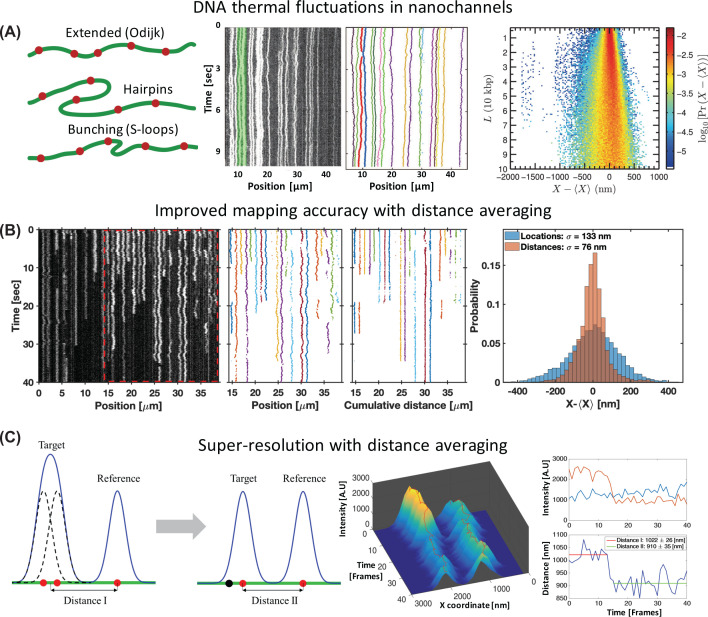
Figure 3. Physical aspects of genomic optical mapping in nanochannels.
(A) DNA fluctuations in nanochannels. Left, possible conformations of DNA molecules confined to nanochannels that contribute to the measured fluctuations. Reprinted from [47], with the permission of AIP Publishing. Middle, raw kymograph (left) and the corresponding marker localizations (right) of barcode labeled genomic DNA extracted from E. coli, showing the thermal fluctuations of confined DNA. Adapted from [48], with the permission of AIP Publishing. Right, probability distribution of observed marker separations in relation to their aligned genomic distance (calculated using 70,305 observed marker positions from single-frame images of 4557 DNA molecules confined to 43 × 43 nm² cross-section channels). The distribution gets wider as the genomic distance increases and therefore mapping accuracy reduces with genomic marker separation. Reprinted from [47], with the permission of AIP Publishing. (B) Improving mapping accuracy with pairwise distance averaging. Left, raw kymograph of the D4Z4 tandem repeat region in chromosome 4 (see optical genome mapping impact section), confined to 45 × 45 nm² cross-section channels (repetitive region marked with the red dashed frame). The repetitive region was used by Jeffet et al. [45] as a ruler to quantify the accuracy and resolution of optical mapping. Adjacent spots in the repetitive segment are distanced 3.3 kbp and each spot is composed of two unresolved fluorophores spaced 676 bp apart. Middle, comparison between localizations maps and cumulative pairwise distances maps of the repetitive region. The localizations map shows correlations in marker fluctuations at short genomic separations, while the distances map shows reduced fluctuations. Right, observed locations distribution compared to the pairwise distance distribution of the ruler molecule’s markers shown to the left. The distances standard deviation is reduced by ∼2-fold compared with locations, allowing increased mapping accuracy. (C) Enabling super-resolution with distance averaging. Left, schematic illustration of the method. A target consisting of multiple sub-diffraction-limit spaced fluorophores is imaged as a single gaussian signal on the camera. Pairwise distance recording between the target and an adjacent marker allows to remove the local collective fluctuations, and thus enable sensitivity to distance shifts originating from the target’s fluorophores blinking or bleaching behavior. Middle, 3D visualization of the intensity–time profile of two fluorescent spots presented in panel (B). Each spot is composed of two fluorophores, where a bleaching event of one of the fluorophores of the left spot is evident after 15 frames. Top right, Intensity time trace of the spots. Bottom right, pairwise distance time-trace between the two spots. The bleaching step alters the mean distance between the two spots allowing to resolve the sub-diffraction-limit distance between them with ∼30 nm resolution. Panels (B and C) were adapted with permission from [45]. Copyright (2016) American Chemical Society.