Fig. 2 |. Mechanical reinforcement and reversible shape transformation of tissue specimens via ELASTicization.
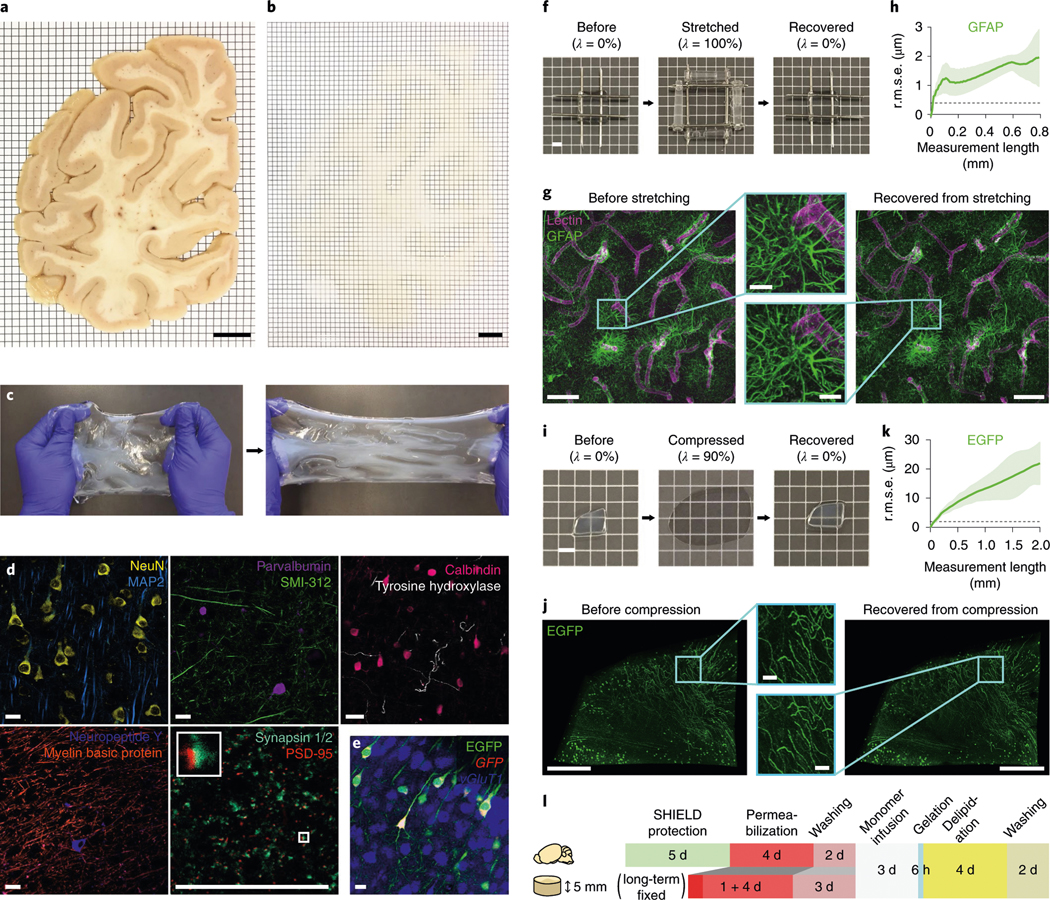
a,b, A 2-mm-thick coronal human brain hemisphere slab from the occipital pole before (a) and after (b) ELAST transformation and RI-matching. Scale bars, 1 cm. Experiment was repeated two times with similar results. c, Stretchability of an ELASTicized human brain slab. d, Immunolabeling of various proteins as indicated in ELASTicized 200-μm-thick human brain cortical samples. Scale bars, 40 μm. Experiments in c and d were repeated two times with similar results. e, In situ hybridization of GFP and vGluT1 mRNAs in an ELASTicized mouse hippocampus expressing EGFP. Scale bar, 40 μm. f, Reversible shape transformation of an ELASTicized 1-mm-thick human brain sample upon twofold stretching in both lateral dimensions. λ, tensile strain. Scale bar, 2 mm. g, Axially projected microscopic images of a prelabeled sample before stretching and after recovery (a representative image from n = 5 samples). Scale bars, 100 μm (insets, 20 μm). h, A mean ± s.d. plot of three-dimensional (3D) geometric deformation measured by the altered distances between feature correspondences in GFAP images at different length scales (n = 5 samples). Dashed line indicates the lateral pixel size of images. r.m.s.e., root mean squared error. i, Reversible shape transformation of a 1.4-mm-thick ELASTicized EGFP-expressing mouse brain sample upon ninefold compression. λ, compression strain. Scale bar, 2 mm. j, Three-dimensionally rendered microscopic images of the sample in i with RI-matching (a representative image from n = 5 samples). Scale bars, 500 μm (insets, 50 μm). k, 3D geometric deformation quantified from EGFP images (n = 5 samples), the same as in h. l, A diagram of the sample-processing procedure for ELASTicization with a suggested timeline. Upper row is for freshly perfused mouse organs, including the brain. Lower row is for about 5-mm-thick specimens from archived human brains that are typically fixed in a formalin solution for a long time. The optimal durations could vary by the degree of fixation and the sample-to-solution volume ratio.
